Cell-free DNA donor fraction analysis in pediatric and adult heart transplant patients by multiplexed allele-specific quantitative PCR: Validation of a rapid and highly sensitive clinical test for stratification of rejection probability
- PMID: 31929557
- PMCID: PMC6957190
- DOI: 10.1371/journal.pone.0227385
Cell-free DNA donor fraction analysis in pediatric and adult heart transplant patients by multiplexed allele-specific quantitative PCR: Validation of a rapid and highly sensitive clinical test for stratification of rejection probability
Abstract
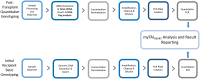
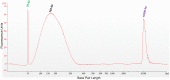
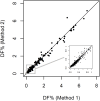
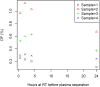
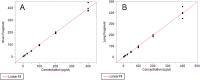
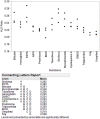
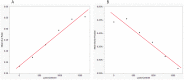
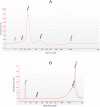
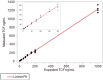
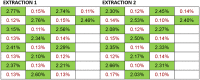
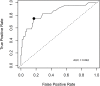
Lifelong noninvasive rejection monitoring in heart transplant patients is a critical clinical need historically poorly met in adults and unavailable for children and infants. Cell-free DNA (cfDNA) donor-specific fraction (DF), a direct marker of selective donor organ injury, is a promising analytical target. Methodological differences in sample processing and DF determination profoundly affect quality and sensitivity of cfDNA analyses, requiring specialized optimization for low cfDNA levels typical of transplant patients. Using next-generation sequencing, we previously correlated elevated DF with acute cellular and antibody-mediated rejection (ACR and AMR) in pediatric and adult heart transplant patients. However, next-generation sequencing is limited by cost, TAT, and sensitivity, leading us to clinically validate a rapid, highly sensitive, quantitative genotyping test, myTAIHEART®, addressing these limitations. To assure pre-analytical quality and consider interrelated cfDNA measures, plasma preparation was optimized and total cfDNA (TCF) concentration, DNA fragmentation, and DF quantification were validated in parallel for integration into myTAIHEART reporting. Analytical validations employed individual and reconstructed mixtures of human blood-derived genomic DNA (gDNA), cfDNA, and gDNA sheared to apoptotic length. Precision, linearity, and limits of blank/detection/quantification were established for TCF concentration, DNA fragmentation ratio, and DF determinations. For DF, multiplexed high-fidelity amplification followed by quantitative genotyping of 94 SNP targets was applied to 1168 samples to evaluate donor options in staged simulations, demonstrating DF call equivalency with/without donor genotype. Clinical validation studies using 158 matched endomyocardial biopsy-plasma pairs from 76 pediatric and adult heart transplant recipients selected a DF cutoff (0.32%) producing 100% NPV for ≥2R ACR. This supports the assay's conservative intended use of stratifying low versus increased probability of ≥2R ACR. myTAIHEART is clinically validated for heart transplant recipients ≥2 months old and ≥8 days post-transplant, expanding opportunity for noninvasive transplant rejection assessment to infants and children and to all recipients >1 week post-transplant.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: PEN, MEM, and AT-M are full time employees and Professors of the Medical College of Wisconsin. PEN is Medical Director of the TAI Diagnostics Clinical Reference Laboratory and its Chief Laboratory Officer. MEM and AT-M are Co-Founders of TAI Diagnostics, Inc. PEN, MEM, AT-M, EZ, DKM, KDS, AT, PD, MAB, AV, MG, HL and CR have stock options at TAI Diagnostics. Multiple relevant patent applications from which authors could benefit are pending. Those patent numbers are PCT/US2013/037439, PCT/US2016/030313, PCT/US2017/030291, PCT/US2017/030292, PCT/US2017/030293, PCT/US2017/059808, PCT/US2017/059802, PCT/US2019/031860, PCT/US2018/038598, PCT/US2018/038604, PCT/US2018/038609, PCT/US2018/000278, and PCT/US2018/065845. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures






Similar articles
-
Donor fraction cell-free DNA and rejection in adult and pediatric heart transplantation.J Heart Lung Transplant. 2020 May;39(5):454-463. doi: 10.1016/j.healun.2019.11.015. Epub 2019 Nov 29. J Heart Lung Transplant. 2020. PMID: 31983667
-
Progress in Noninvasive Surveillance for Acute Rejection in Pediatric Heart Transplant Recipients: A Real-World Analysis of Donor-Derived Cell-Free DNA-Based Surveillance Protocol.Clin Transplant. 2024 Oct;38(10):e15481. doi: 10.1111/ctr.15481. Clin Transplant. 2024. PMID: 39412485
-
Clinical Utility of Donor-Derived Cell-Free DNA in Heart Transplant Recipients With Multi-Organ Transplants.Clin Transplant. 2024 Oct;38(10):e15479. doi: 10.1111/ctr.15479. Clin Transplant. 2024. PMID: 39422086
-
Donor-specific Cell-free DNA as a Biomarker in Solid Organ Transplantation. A Systematic Review.Transplantation. 2019 Feb;103(2):273-283. doi: 10.1097/TP.0000000000002482. Transplantation. 2019. PMID: 30308576
-
The Monitoring of Donor-derived Cell-free DNA in Kidney Transplantation.Transplantation. 2021 Mar 1;105(3):509-516. doi: 10.1097/TP.0000000000003393. Transplantation. 2021. PMID: 32732615 Review.
Cited by
-
Upregulated circulating mir-424 and its' diagnostic value for gram-negative bacteremia after thoracic transplantation.Noncoding RNA Res. 2022 Aug 31;7(4):217-225. doi: 10.1016/j.ncrna.2022.08.001. eCollection 2022 Dec. Noncoding RNA Res. 2022. PMID: 36187569 Free PMC article.
-
Non-invasive cardiac allograft rejection surveillance: reliability and clinical value for prevention of heart failure.Heart Fail Rev. 2021 Mar;26(2):319-336. doi: 10.1007/s10741-020-10023-3. Epub 2020 Sep 5. Heart Fail Rev. 2021. PMID: 32889634 Review.
-
The Organ Trail: A Review of Biomarkers of Organ Failure.Front Oncol. 2020 Nov 11;10:579219. doi: 10.3389/fonc.2020.579219. eCollection 2020. Front Oncol. 2020. PMID: 33262945 Free PMC article. Review.
-
Free-Circulating Nucleic Acids as Biomarkers in Patients After Solid Organ Transplantation.Ann Transplant. 2023 Aug 15;28:e939750. doi: 10.12659/AOT.939750. Ann Transplant. 2023. PMID: 37580899 Free PMC article. Review.
-
Validation of donor fraction cell-free DNA with biopsy-proven cardiac allograft rejection in children and adults.J Thorac Cardiovasc Surg. 2023 Feb;165(2):460-468.e2. doi: 10.1016/j.jtcvs.2022.04.027. Epub 2022 Apr 30. J Thorac Cardiovasc Surg. 2023. PMID: 35643770 Free PMC article.
References
-
- Mena C, Wencker D, Krumholz HM, McNamara RL. Detection of heart transplant rejection in adults by echocardiographic diastolic indices: a systematic review of the literature. Journal of the American Society of Echocardiography: official publication of the American Society of Echocardiography. 2006;19(10):1295–300 - PubMed
-
- Vermes E, Pantaleon C, Auvet A, Cazeneuve N, Machet MC, Delhommais A, et al. Cardiovascular magnetic resonance in heart transplant patients: diagnostic value of quantitative tissue markers: T2 mapping and extracellular volume fraction, for acute rejection diagnosis. Journal of cardiovascular magnetic resonance: official journal of the Society for Cardiovascular Magnetic Resonance. 2018;20(1):59. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

