Genome ancestry mosaics reveal multiple and cryptic contributors to cultivated banana
- PMID: 31930580
- PMCID: PMC7317953
- DOI: 10.1111/tpj.14683
Genome ancestry mosaics reveal multiple and cryptic contributors to cultivated banana
Abstract
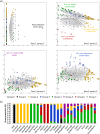
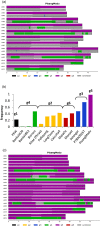
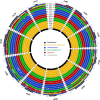
Hybridizations between closely related species commonly occur in the domestication process of many crops. Banana cultivars are derived from such hybridizations between species and subspecies of the Musa genus that have diverged in various tropical Southeast Asian regions and archipelagos. Among the diploid and triploid hybrids generated, those with seedless parthenocarpic fruits were selected by humans and thereafter dispersed through vegetative propagation. Musa acuminata subspecies contribute to most of these cultivars. We analyzed sequence data from 14 M. acuminata wild accessions and 10 M. acuminata-based cultivars, including diploids and one triploid, to characterize the ancestral origins along their chromosomes. We used multivariate analysis and single nucleotide polymorphism clustering and identified five ancestral groups as contributors to these cultivars. Four of these corresponded to known M. acuminata subspecies. A fifth group, found only in cultivars, was defined based on the 'Pisang Madu' cultivar and represented two uncharacterized genetic pools. Diverse ancestral contributions along cultivar chromosomes were found, resulting in mosaics with at least three and up to five ancestries. The commercially important triploid Cavendish banana cultivar had contributions from at least one of the uncharacterized genetic pools and three known M. acuminata subspecies. Our results highlighted that cultivated banana origins are more complex than expected - involving multiple hybridization steps - and also that major wild banana ancestors have yet to be identified. This study revealed the extent to which admixture has framed the evolution and domestication of a crop plant.
Keywords: Musa acuminata; admixture; diversity; genome ancestry; hybridization.
© 2020 The Authors. The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Boonruangrod, R. , Desai, D. , Fluch, S. , Berenyi, M. and Burg, K. (2008) Identification of cytoplasmic ancestor gene‐pools of Musa acuminata Colla and Musa balbisiana Colla and their hybrids by chloroplast and mitochondrial haplotyping. Theor. Appl. Genet. 118, 43–55. - PubMed
-
- Bredeson, J.V. , Lyons, J.B. , Prochnik, S.E. et al . (2016) Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nat. Biotechnol. 34, 562–570. - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources

