Post-translational modifications of Drosophila melanogaster HOX protein, Sex combs reduced
- PMID: 31931520
- PMCID: PMC6957346
- DOI: 10.1371/journal.pone.0227642
Post-translational modifications of Drosophila melanogaster HOX protein, Sex combs reduced
Abstract
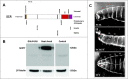
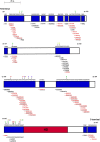
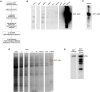
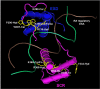
Homeotic selector (HOX) transcription factors (TFs) regulate gene expression that determines the identity of Drosophila segments along the anterior-posterior (A-P) axis. The current challenge with HOX proteins is understanding how they achieve their functional specificity while sharing a highly conserved homeodomain (HD) that recognize the same DNA binding sites. One mechanism proposed to regulate HOX activity is differential post-translational modification (PTM). As a first step in investigating this hypothesis, the sites of PTM on a Sex combs reduced protein fused to a triple tag (SCRTT) extracted from developing embryos were identified by Tandem Mass Spectrometry (MS/MS). The PTMs identified include phosphorylation at S185, S201, T315, S316, T317 and T324, acetylation at K218, S223, S227, K309, K434 and K439, formylation at K218, K309, K325, K341, K369, K434 and K439, methylation at S19, S166, K168 and T364, carboxylation at D108, K298, W307, K309, E323, K325 and K369, and hydroxylation at P22, Y87, P107, D108, D111, P269, P306, R310, N321, K325, Y334, R366, P392 and Y398. Of the 44 modifications, 18 map to functionally important regions of SCR. Besides a highly conserved DNA-binding HD, HOX proteins also have functionally important, evolutionarily conserved small motifs, which may be Short Linear Motifs (SLiMs). SLiMs are proposed to be preferential sites of phosphorylation. Although 6 of 7 phosphosites map to regions of predicted SLiMs, we find no support for the hypothesis that the individual S, T and Y residues of predicted SLiMs are phosphorylated more frequently than S, T and Y residues outside of predicted SLiMs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
A Distalless-responsive enhancer of the Hox gene Sex combs reduced is required for segment- and sex-specific sensory organ development in Drosophila.PLoS Genet. 2018 Apr 10;14(4):e1007320. doi: 10.1371/journal.pgen.1007320. eCollection 2018 Apr. PLoS Genet. 2018. PMID: 29634724 Free PMC article.
-
Dissecting the functional specificities of two Hox proteins.Genes Dev. 2010 Jul 15;24(14):1533-45. doi: 10.1101/gad.1936910. Genes Dev. 2010. PMID: 20634319 Free PMC article.
-
Inhibitory activities of short linear motifs underlie Hox interactome specificity in vivo.Elife. 2015 Apr 14;4:e06034. doi: 10.7554/eLife.06034. Elife. 2015. PMID: 25869471 Free PMC article.
-
Differential pleiotropy and HOX functional organization.Dev Biol. 2015 Feb 1;398(1):1-10. doi: 10.1016/j.ydbio.2014.11.001. Epub 2014 Nov 11. Dev Biol. 2015. PMID: 25448696 Review.
-
Lessons on gene regulation learnt from the Drosophila melanogaster bithorax complex.Int J Dev Biol. 2020;64(1-2-3):151-158. doi: 10.1387/ijdb.190167rm. Int J Dev Biol. 2020. PMID: 32659003 Review.
Cited by
-
Versatile roles of disordered transcription factor effector domains in transcriptional regulation.FEBS J. 2025 Jun;292(12):3014-3033. doi: 10.1111/febs.17424. Epub 2025 Jan 30. FEBS J. 2025. PMID: 39888268 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

