Cytokine mRNA Degradation in Cardiomyocytes Restrains Sterile Inflammation in Pressure-Overloaded Hearts
- PMID: 31931613
- PMCID: PMC7034406
- DOI: 10.1161/CIRCULATIONAHA.119.044582
Cytokine mRNA Degradation in Cardiomyocytes Restrains Sterile Inflammation in Pressure-Overloaded Hearts
Erratum in
-
Correction to: Cytokine mRNA Degradation in Cardiomyocytes Restrains Sterile Inflammation in Pressure-Overloaded Hearts.Circulation. 2024 Apr 16;149(16):e1131. doi: 10.1161/CIR.0000000000001248. Epub 2024 Apr 15. Circulation. 2024. PMID: 38620089 Free PMC article. No abstract available.
Abstract
Background: Proinflammatory cytokines play an important role in the pathogenesis of heart failure. The mechanisms responsible for maintaining sterile inflammation within failing hearts remain poorly defined. Although transcriptional control is important for proinflammatory cytokine gene expression, the stability of mRNA also contributes to the kinetics of immune responses. Regnase-1 is an RNase involved in the degradation of a set of proinflammatory cytokine mRNAs in immune cells. The role of Regnase-1 in nonimmune cells such as cardiomyocytes remains to be elucidated.
Methods: To examine the role of proinflammatory cytokine degradation by Regnase-1 in cardiomyocytes, cardiomyocyte-specific Regnase-1-deficient mice were generated. The mice were subjected to pressure overload by means of transverse aortic constriction to induce heart failure. Cardiac remodeling was assessed by echocardiography as well as histological and molecular analyses 4 weeks after operation. Inflammatory cell infiltration was examined by immunostaining. Interleukin-6 signaling was inhibited by administration with its receptor antibody. Overexpression of Regnase-1 in the heart was performed by adeno-associated viral vector-mediated gene transfer.
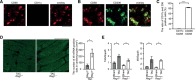
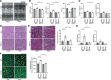
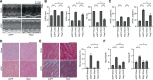
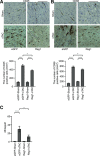
Results: Cardiomyocyte-specific Regnase-1-deficient mice showed no cardiac phenotypes under baseline conditions, but exhibited severe inflammation and dilated cardiomyopathy after 4 weeks of pressure overload compared with control littermates. Four weeks after transverse aortic constriction, the Il6 mRNA level was upregulated, but not other cytokine mRNAs, including tumor necrosis factor-α, in Regnase-1-deficient hearts. Although the Il6 mRNA level increased 1 week after operation in both Regnase-1-deficient and control hearts, it showed no increase in control hearts 4 weeks after operation. Administration of anti-interleukin-6 receptor antibody attenuated the development of inflammation and cardiomyopathy in cardiomyocyte-specific Regnase-1-deficient mice. In severe pressure overloaded wild-type mouse hearts, sustained induction of Il6 mRNA was observed, even though the protein level of Regnase-1 increased. Adeno-associated virus 9-mediated cardiomyocyte-targeted gene delivery of Regnase-1 or administration of anti-interleukin-6 receptor antibody attenuated the development of cardiomyopathy induced by severe pressure overload in wild-type mice.
Conclusions: The degradation of cytokine mRNA by Regnase-1 in cardiomyocytes plays an important role in restraining sterile inflammation in failing hearts and the Regnase-1-mediated pathway might be a therapeutic target to treat patients with heart failure.
Keywords: RNA stability; heart failure; inflammation; interleukin-6.
Figures



References
-
- Mann DL, McMurray JJ, Packer M, Swedberg K, Borer JS, Colucci WS, Djian J, Drexler H, Feldman A, Kober L, et al. Targeted anticytokine therapy in patients with chronic heart failure: results of the Randomized Etanercept Worldwide Evaluation (RENEWAL). Circulation. 2004;109:1594–1602. doi: 10.1161/01.CIR.0000124490.27666.B2. - PubMed
-
- Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

