High-brightness anterograde transneuronal HSV1 H129 tracer modified using a Trojan horse-like strategy
- PMID: 31931837
- PMCID: PMC6958791
- DOI: 10.1186/s13041-020-0544-2
High-brightness anterograde transneuronal HSV1 H129 tracer modified using a Trojan horse-like strategy
Abstract
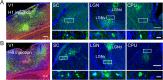
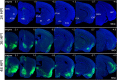
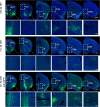
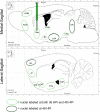
Neurotropic viral transsynaptic tracing is an increasingly powerful technique for dissecting the structure and function of neural circuits. Herpes simplex virus type 1 strain H129 has been widely used as an anterograde tracer. However, HSV tracers still have several shortcomings, including high toxicity, low sensitivity and non-specific retrograde labeling. Here, we aimed to construct high-brightness HSV anterograde tracers by increasing the expression of exogenous genes carried by H129 viruses. Using a Trojan horse-like strategy, a HSV/AAV (adeno-associated virus) chimaera termed H8 was generated to enhance the expression of a fluorescent marker. In vitro and in vivo assays showed that the exogenous gene was efficiently replicated and amplified by the synergism of the HSV vector and introduced AAV replication system. H8 reporting fluorescence was brighter than that of currently available H129 tracers, and H8 could be used for fast and effective anterograde tracing without additional immunostaining. These results indicated that foreign gene expression in HSV tracers could be enhanced by integrating HSV with AAV replication system. This approach may be useful as a general enhanced expression strategy for HSV-based tracing tools or gene delivery vectors.
Keywords: Adeno-associated virus; Anterograde tracer; H129; Herpes simplex virus; High-brightness; Neural circuit tracing; Trojan horse-like strategy.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
A novel H129-based anterograde monosynaptic tracer exhibits features of strong labeling intensity, high tracing efficiency, and reduced retrograde labeling.Mol Neurodegener. 2022 Jan 10;17(1):6. doi: 10.1186/s13024-021-00508-6. Mol Neurodegener. 2022. PMID: 35012591 Free PMC article.
-
Anterograde Viral Tracer Herpes Simplex Virus 1 Strain H129 Transports Primarily as Capsids in Cortical Neuron Axons.J Virol. 2020 Mar 31;94(8):e01957-19. doi: 10.1128/JVI.01957-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31969440 Free PMC article.
-
The neuroinvasive profiles of H129 (herpes simplex virus type 1) recombinants with putative anterograde-only transneuronal spread properties.Brain Struct Funct. 2015;220(3):1395-420. doi: 10.1007/s00429-014-0733-9. Epub 2014 Mar 2. Brain Struct Funct. 2015. PMID: 24585022 Free PMC article.
-
Anterograde Neuronal Circuit Tracers Derived from Herpes Simplex Virus 1: Development, Application, and Perspectives.Int J Mol Sci. 2020 Aug 18;21(16):5937. doi: 10.3390/ijms21165937. Int J Mol Sci. 2020. PMID: 32824837 Free PMC article. Review.
-
Herpes simplex virus type 1-based amplicon vector systems.Adv Virus Res. 2000;55:425-51. doi: 10.1016/s0065-3527(00)55011-8. Adv Virus Res. 2000. PMID: 11050950 Review. No abstract available.
Cited by
-
Neuropeptide S Attenuates the Alarm Pheromone-Evoked Defensive and Risk Assessment Behaviors Through Activation of Cognate Receptor-Expressing Neurons in the Posterior Medial Amygdala.Front Mol Neurosci. 2021 Dec 24;14:752516. doi: 10.3389/fnmol.2021.752516. eCollection 2021. Front Mol Neurosci. 2021. PMID: 35002616 Free PMC article.
-
Brain-wide neuronal circuit connectome of human glioblastoma.Nature. 2025 May;641(8061):222-231. doi: 10.1038/s41586-025-08634-7. Epub 2025 Jan 16. Nature. 2025. PMID: 39821165 Free PMC article.
-
A new anterograde trans-synaptic tracer based on Sindbis virus.Neural Regen Res. 2022 Dec;17(12):2761-2764. doi: 10.4103/1673-5374.339495. Neural Regen Res. 2022. PMID: 35662226 Free PMC article.
-
A novel H129-based anterograde monosynaptic tracer exhibits features of strong labeling intensity, high tracing efficiency, and reduced retrograde labeling.Mol Neurodegener. 2022 Jan 10;17(1):6. doi: 10.1186/s13024-021-00508-6. Mol Neurodegener. 2022. PMID: 35012591 Free PMC article.
-
The connectome from the cerebral cortex to skeletal muscle using viral transneuronal tracers: a review.Am J Transl Res. 2022 Jul 15;14(7):4864-4879. eCollection 2022. Am J Transl Res. 2022. PMID: 35958450 Free PMC article. Review.
References
-
- Card JP, Enquist LW. Current protocols in neuroscience. 2001. Transneuronal circuit analysis with pseudorabies viruses. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

