An Ancient, MHC-Linked, Nonclassical Class I Lineage in Cartilaginous Fish
- PMID: 31932500
- PMCID: PMC7002201
- DOI: 10.4049/jimmunol.1901025
An Ancient, MHC-Linked, Nonclassical Class I Lineage in Cartilaginous Fish
Abstract
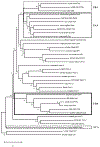
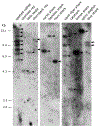
Cartilaginous fishes, or chondrichthyans, are the oldest jawed vertebrates that have an adaptive immune system based on the MHC and Ig superfamily-based AgR. In this basal group of jawed vertebrates, we identified a third nonclassical MHC class I lineage (UDA), which is present in all species analyzed within the two major cartilaginous subclasses, Holocephali (chimaeras) and Elasmobranchii (sharks, skates, and rays). The deduced amino acid sequences of UDA have eight out of nine typically invariant residues that bind to the N and C termini of bound peptide found in most vertebrae classical class I (UAA); additionally, the other predicted 28 peptide-binding residues are perfectly conserved in all elasmobranch UDA sequences. UDA is distinct from UAA in its differential tissue distribution and its lower expression levels and is mono- or oligomorphic unlike the highly polymorphic UAA UDA has a low copy number in elasmobranchs but is multicopy in the holocephalan spotted ratfish (Hydrolagus colliei). Using a nurse shark (Ginglymostoma cirratum) family, we found that UDA is MHC linked but separable by recombination from the tightly linked cluster of UAA, TAP, and LMP genes, the so-called class I region found in most nonmammalian vertebrates. UDA has predicted structural features that are similar to certain nonclassical class I genes in other vertebrates, and, unlike polymorpic classical class I, we anticipate that it may bind to a conserved set of specialized peptides.
Copyright © 2020 by The American Association of Immunologists, Inc.
Figures





Similar articles
-
A Highly Complex, MHC-Linked, 350 Million-Year-Old Shark Nonclassical Class I Lineage.J Immunol. 2021 Aug 1;207(3):824-836. doi: 10.4049/jimmunol.2000851. Epub 2021 Jul 23. J Immunol. 2021. PMID: 34301841 Free PMC article.
-
Proteasome, transporter associated with antigen processing, and class I genes in the nurse shark Ginglymostoma cirratum: evidence for a stable class I region and MHC haplotype lineages.J Immunol. 2002 Jan 15;168(2):771-81. doi: 10.4049/jimmunol.168.2.771. J Immunol. 2002. PMID: 11777971 Free PMC article.
-
Cartilaginous fish class II genes reveal unprecedented old allelic lineages and confirm the late evolutionary emergence of DM.Mol Immunol. 2020 Dec;128:125-138. doi: 10.1016/j.molimm.2020.10.003. Epub 2020 Oct 27. Mol Immunol. 2020. PMID: 33126081 Free PMC article.
-
What sharks can tell us about the evolution of MHC genes.Immunol Rev. 1998 Dec;166:317-31. doi: 10.1111/j.1600-065x.1998.tb01272.x. Immunol Rev. 1998. PMID: 9914922 Review.
-
Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man.Immunol Rev. 2002 Dec;190:95-122. doi: 10.1034/j.1600-065x.2002.19008.x. Immunol Rev. 2002. PMID: 12493009 Review.
Cited by
-
An integrated approach to the characterization of immune repertoires using AIMS: An Automated Immune Molecule Separator.PLoS Comput Biol. 2023 Oct 20;19(10):e1011577. doi: 10.1371/journal.pcbi.1011577. eCollection 2023 Oct. PLoS Comput Biol. 2023. PMID: 37862356 Free PMC article.
-
Allele co-segregation and haplotype diversity of MHC IIβ genes in the small-spotted catshark Scyliorhinus canicula.Immunogenetics. 2025 Mar 31;77(1):19. doi: 10.1007/s00251-025-01376-w. Immunogenetics. 2025. PMID: 40164821 Free PMC article.
-
Extensive MHC class IIβ diversity across multiple loci in the small-spotted catshark (Scyliorhinus canicula).Sci Rep. 2023 Mar 7;13(1):3837. doi: 10.1038/s41598-023-30876-6. Sci Rep. 2023. PMID: 36882519 Free PMC article.
-
An Ancestral Major Histocompatibility Complex Organization in Cartilaginous Fish: Reconstructing MHC Origin and Evolution.Mol Biol Evol. 2023 Dec 1;40(12):msad262. doi: 10.1093/molbev/msad262. Mol Biol Evol. 2023. PMID: 38059517 Free PMC article.
-
γδ T, NKT, and MAIT Cells During Evolution: Redundancy or Specialized Functions?J Immunol. 2022 Jul 15;209(2):217-225. doi: 10.4049/jimmunol.2200105. J Immunol. 2022. PMID: 35821101 Free PMC article. Review.
References
-
- Warr GW 1997. The adaptive immune system of fish. Dev Biol Stand. 90: 15–21. - PubMed
-
- Flajnik MF, and Rumfelt L. 2000. The immune system of cartilaginous fish In: Origin and evolution of the vertebrate immune system. Springer Berlin, Heidelberg; 249–270. - PubMed
-
- Bartl S, Baish MA, Flajnik MF, and Ohta Y. 1997. Identification of class I genes in cartilaginous fish, the most ancient group of vertebrates displaying an adaptive immune response. J Immunol. 159: 6097–6104. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

