Antimicrobial resistant enteric bacteria are widely distributed amongst people, animals and the environment in Tanzania
- PMID: 31932601
- PMCID: PMC6957491
- DOI: 10.1038/s41467-019-13995-5
Antimicrobial resistant enteric bacteria are widely distributed amongst people, animals and the environment in Tanzania
Abstract
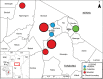
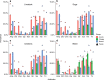
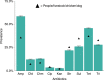
Antibiotic use and bacterial transmission are responsible for the emergence, spread and persistence of antimicrobial-resistant (AR) bacteria, but their relative contribution likely differs across varying socio-economic, cultural, and ecological contexts. To better understand this interaction in a multi-cultural and resource-limited context, we examine the distribution of antimicrobial-resistant enteric bacteria from three ethnic groups in Tanzania. Household-level data (n = 425) was collected and bacteria isolated from people, livestock, dogs, wildlife and water sources (n = 62,376 isolates). The relative prevalence of different resistance phenotypes is similar across all sources. Multi-locus tandem repeat analysis (n = 719) and whole-genome sequencing (n = 816) of Escherichia coli demonstrate no evidence for host-population subdivision. Multivariate models show no evidence that veterinary antibiotic use increased the odds of detecting AR bacteria, whereas there is a strong association with livelihood factors related to bacterial transmission, demonstrating that to be effective, interventions need to accommodate different cultural practices and resource limitations.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- World Health Organization. Global action plan on antimicrobial resistance. (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

