The interaction of Multiple Sclerosis risk loci with Epstein-Barr virus phenotypes implicates the virus in pathogenesis
- PMID: 31932685
- PMCID: PMC6957475
- DOI: 10.1038/s41598-019-55850-z
The interaction of Multiple Sclerosis risk loci with Epstein-Barr virus phenotypes implicates the virus in pathogenesis
Abstract
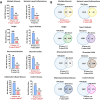
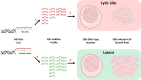
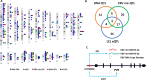
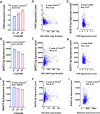
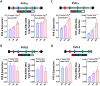
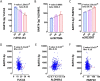
Translating the findings of genome wide association studies (GWAS) to new therapies requires identification of the relevant immunological contexts to interrogate for genetic effects. In one of the largest GWAS, more than 200 risk loci have been identified for Multiple Sclerosis (MS) susceptibility. Infection with Epstein-Barr virus (EBV) appears to be necessary for the development of Multiple Sclerosis (MS). Many MS risk loci are associated with altered gene expression in EBV infected B cells (LCLs). We have interrogated this immunological context to identify interaction between MS risk loci and EBV DNA copy number, intrinsic growth rate and EBV encoded miRNA expression. The EBV DNA copy number was associated with significantly more risk alleles for MS than for other diseases or traits. EBV miRNAs BART4-3p and BART3-5p were highly associated with EBV DNA copy number and MS risk loci. The poliovirus receptor (PVR) risk SNP was associated with EBV DNA copy number, PVR and miRNA expression. Targeting EBV miRNAs BART4-3p and BART3-5p, and the gene PVR, may provide therapeutic benefit in MS. This study also indicates how immunological context and risk loci interactions can be exploited to validate and develop novel therapeutic approaches.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Ryan Katie J., White Charles C., Patel Kruti, Xu Jishu, Olah Marta, Replogle Joseph M., Frangieh Michael, Cimpean Maria, Winn Phoebe, McHenry Allison, Kaskow Belinda J., Chan Gail, Cuerdon Nicole, Bennett David A., Boyd Justin D., Imitola Jaime, Elyaman Wassim, De Jager Philip L., Bradshaw Elizabeth M. A human microglia-like cellular model for assessing the effects of neurodegenerative disease gene variants. Science Translational Medicine. 2017;9(421):eaai7635. doi: 10.1126/scitranslmed.aai7635. - DOI - PMC - PubMed
-
- Patsopoulos, N. et al. The Multiple Sclerosis Genomic Map: Role of peripheral immune cells and resident microglia in susceptibility. bioRxiv, 10.1101/143933 (2017).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

