FLOW-MAP: a graph-based, force-directed layout algorithm for trajectory mapping in single-cell time course datasets
- PMID: 31932774
- PMCID: PMC7897424
- DOI: 10.1038/s41596-019-0246-3
FLOW-MAP: a graph-based, force-directed layout algorithm for trajectory mapping in single-cell time course datasets
Abstract
High-dimensional single-cell technologies present new opportunities for biological discovery, but the complex nature of the resulting datasets makes it challenging to perform comprehensive analysis. One particular challenge is the analysis of single-cell time course datasets: how to identify unique cell populations and track how they change across time points. To facilitate this analysis, we developed FLOW-MAP, a graphical user interface (GUI)-based software tool that uses graph layout analysis with sequential time ordering to visualize cellular trajectories in high-dimensional single-cell datasets obtained from flow cytometry, mass cytometry or single-cell RNA sequencing (scRNAseq) experiments. Here we provide a detailed description of the FLOW-MAP algorithm and how to use the open-source R package FLOWMAPR via its GUI or with text-based commands. This approach can be applied to many dynamic processes, including in vitro stem cell differentiation, in vivo development, oncogenesis, the emergence of drug resistance and cell signaling dynamics. To demonstrate our approach, we perform a step-by-step analysis of a single-cell mass cytometry time course dataset from mouse embryonic stem cells differentiating into the three germ layers: endoderm, mesoderm and ectoderm. In addition, we demonstrate FLOW-MAP analysis of a previously published scRNAseq dataset. Using both synthetic and experimental datasets for comparison, we perform FLOW-MAP analysis side by side with other single-cell analysis methods, to illustrate when it is advantageous to use the FLOW-MAP approach. The protocol takes between 30 min and 1.5 h to complete.
Conflict of interest statement
Competing interests
G.P.N. is a paid consultant for Fluidigm, the manufacturer that produced some of the reagents and instrumentation used in this study. The remaining authors declare no competing interests.
Figures




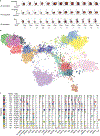

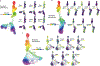

References
-
- Jolliffe IT Principal Component Analysis (Springer-Verlag, 2002).
-
- Ringnér M What is principal component analysis? Nat. Biotechnol 26, 303–304 (2008). - PubMed
-
- van der Maaten L & Hinton G Visualizing data using t-SNE. J. Mach. Learn. Res 9, 2579–2605 (2008).
Publication types
MeSH terms
Grants and funding
- R01CA184968/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)/International
- 5T32LM012416/U.S. Department of Health & Human Services | NIH | U.S. National Library of Medicine (NLM)/International
- R33 CA183654/CA/NCI NIH HHS/United States
- 1R33CA183654-01/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)/International
- 5UH2AR067676/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)/International
- U19 AI100627/AI/NIAID NIH HHS/United States
- T32 LM012416/LM/NLM NIH HHS/United States
- 5T32HL007284/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)/International
- R33CA183692/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)/International
- R21 CA183660/CA/NCI NIH HHS/United States
- 1R01NS08953304/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)/International
- U19 AI057229/AI/NIAID NIH HHS/United States
- 1R01GM10983601/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)/International
- F32 GM093508/GM/NIGMS NIH HHS/United States
- 1R21CA183660/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)/International
- R01 HL120724/HL/NHLBI NIH HHS/United States
- T32 GM007276/GM/NIGMS NIH HHS/United States
- T32GM007276/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)/International
- T32 GM008715/GM/NIGMS NIH HHS/United States
- 1U19AI100627/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)/International
- R33 CA183692/CA/NCI NIH HHS/United States
- T32 HL007284/HL/NHLBI NIH HHS/United States
- 5T32GM008715/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)/International
- R01 CA184968/CA/NCI NIH HHS/United States
- UH2 AR067676/AR/NIAMS NIH HHS/United States
- R01HL120724/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)/International

