Influenza-induced monocyte-derived alveolar macrophages confer prolonged antibacterial protection
- PMID: 31932810
- PMCID: PMC6983324
- DOI: 10.1038/s41590-019-0568-x
Influenza-induced monocyte-derived alveolar macrophages confer prolonged antibacterial protection
Abstract
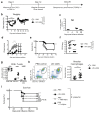
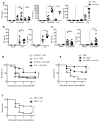
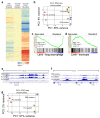
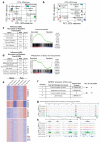
Despite the prevalence and clinical importance of influenza, its long-term effect on lung immunity is unclear. Here we describe that following viral clearance and clinical recovery, at 1 month after infection with influenza, mice are better protected from Streptococcus pneumoniae infection due to a population of monocyte-derived alveolar macrophages (AMs) that produce increased interleukin-6. Influenza-induced monocyte-derived AMs have a surface phenotype similar to resident AMs but display a unique functional, transcriptional and epigenetic profile that is distinct from resident AMs. In contrast, influenza-experienced resident AMs remain largely similar to naive AMs. Thus, influenza changes the composition of the AM population to provide prolonged antibacterial protection. Monocyte-derived AMs persist over time but lose their protective profile. Our results help to understand how transient respiratory infections, a common occurrence in human life, can constantly alter lung immunity by contributing monocyte-derived, recruited cells to the AM population.
Conflict of interest statement
E.M.H. and S.B. were employees of GSK at the time of this study. The other authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

