Multivariate patterning of human pluripotent cells under perfusion reveals critical roles of induced paracrine factors in kidney organoid development
- PMID: 31934619
- PMCID: PMC6949035
- DOI: 10.1126/sciadv.aaw2746
Multivariate patterning of human pluripotent cells under perfusion reveals critical roles of induced paracrine factors in kidney organoid development
Abstract
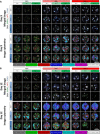
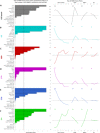
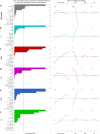
Creating complex multicellular kidney organoids from pluripotent stem cells shows great promise. Further improvements in differentiation outcomes, patterning, and maturation of specific cell types are, however, intrinsically limited by standard tissue culture approaches. We describe a novel full factorial microbioreactor array-based methodology to achieve rapid interrogation and optimization of this complex multicellular differentiation process in a facile manner. We successfully recapitulate early kidney tissue patterning events, exploring more than 1000 unique conditions in an unbiased and quantitative manner, and define new media combinations that achieve near-pure renal cell type specification. Single-cell resolution identification of distinct renal cell types within multilayered kidney organoids, coupled with multivariate analysis, defined the definitive roles of Wnt, fibroblast growth factor, and bone morphogenetic protein signaling in their specification, exposed retinoic acid as a minimal effector of nephron patterning, and highlighted critical contributions of induced paracrine factors on cell specification and patterning.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures



Similar articles
-
Microbioreactor arrays for full factorial screening of exogenous and paracrine factors in human embryonic stem cell differentiation.PLoS One. 2012;7(12):e52405. doi: 10.1371/journal.pone.0052405. Epub 2012 Dec 26. PLoS One. 2012. PMID: 23300662 Free PMC article.
-
Generation of nephron progenitor cells and kidney organoids from human pluripotent stem cells.Nat Protoc. 2017 Jan;12(1):195-207. doi: 10.1038/nprot.2016.170. Epub 2016 Dec 22. Nat Protoc. 2017. PMID: 28005067 Free PMC article.
-
Generation of human antral and fundic gastric organoids from pluripotent stem cells.Nat Protoc. 2019 Jan;14(1):28-50. doi: 10.1038/s41596-018-0080-z. Nat Protoc. 2019. PMID: 30470820 Free PMC article.
-
Pluripotent stem cell-derived kidney organoids: An in vivo-like in vitro technology.Eur J Pharmacol. 2016 Nov 5;790:12-20. doi: 10.1016/j.ejphar.2016.06.059. Epub 2016 Jul 1. Eur J Pharmacol. 2016. PMID: 27375081 Review.
-
Dissecting nephron morphogenesis using kidney organoids from human pluripotent stem cells.Curr Opin Genet Dev. 2022 Feb;72:22-29. doi: 10.1016/j.gde.2021.10.002. Epub 2021 Nov 12. Curr Opin Genet Dev. 2022. PMID: 34781071 Review.
Cited by
-
Critical Analysis of cGMP Large-Scale Expansion Process in Bioreactors of Human Induced Pluripotent Stem Cells in the Framework of Quality by Design.BioDrugs. 2021 Nov;35(6):693-714. doi: 10.1007/s40259-021-00503-9. Epub 2021 Nov 2. BioDrugs. 2021. PMID: 34727354 Free PMC article. Review.
-
ECM-derived biomaterials for regulating tissue multicellularity and maturation.iScience. 2024 Feb 6;27(3):109141. doi: 10.1016/j.isci.2024.109141. eCollection 2024 Mar 15. iScience. 2024. PMID: 38405613 Free PMC article. Review.
-
Dynamic Kidney Organoid Microphysiological Analysis Platform.bioRxiv [Preprint]. 2024 Oct 29:2024.10.27.620552. doi: 10.1101/2024.10.27.620552. bioRxiv. 2024. PMID: 39554191 Free PMC article. Preprint.
-
The "3Ds" of Growing Kidney Organoids: Advances in Nephron Development, Disease Modeling, and Drug Screening.Cells. 2023 Feb 8;12(4):549. doi: 10.3390/cells12040549. Cells. 2023. PMID: 36831216 Free PMC article. Review.
-
Modelling ciliopathy phenotypes in human tissues derived from pluripotent stem cells with genetically ablated cilia.Nat Biomed Eng. 2022 Apr;6(4):463-475. doi: 10.1038/s41551-022-00880-8. Epub 2022 Apr 27. Nat Biomed Eng. 2022. PMID: 35478224 Free PMC article.
References
-
- Takasato M., Er P. X., Becroft M., Vanslambrouck J. M., Stanley E. G., Elefanty A. G., Little M. H., Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat. Cell Biol. 16, 118–126 (2014). - PubMed
-
- Takasato M., Little M. H., The origin of the mammalian kidney: Implications for recreating the kidney in vitro. Development 142, 1937–1947 (2015). - PubMed
-
- Takasato M., Er P. X., Chiu H. S., Maier B., Baillie G. J., Ferguson C., Parton R. G., Wolvetang E. J., Roost M. S., Chuva de Sousa Lopes S. M., Little M. H., Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015). - PubMed
-
- Xia Y., Nivet E., Sancho-Martinez I., Gallegos T., Suzuki K., Okamura D., Wu M.-Z., Dubova I., Esteban C. R., Montserrat N., Campistol J. M., Izpisua Belmonte J. C., Directed differentiation of human pluripotent cells to ureteric bud kidney progenitor-like cells. Nat. Cell Biol. 15, 1507–1515 (2013). - PubMed
-
- Taguchi A., Kaku Y., Ohmori T., Sharmin S., Ogawa M., Sasaki H., Nishinakamura R., Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell 14, 53–67 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

