Disordered chromatin packing regulates phenotypic plasticity
- PMID: 31934628
- PMCID: PMC6949045
- DOI: 10.1126/sciadv.aax6232
Disordered chromatin packing regulates phenotypic plasticity
Abstract
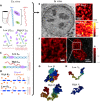
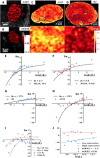
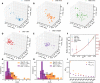
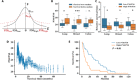
Three-dimensional supranucleosomal chromatin packing plays a profound role in modulating gene expression by regulating transcription reactions through mechanisms such as gene accessibility, binding affinities, and molecular diffusion. Here, we use a computational model that integrates disordered chromatin packing (CP) with local macromolecular crowding (MC) to study how physical factors, including chromatin density, the scaling of chromatin packing, and the size of chromatin packing domains, influence gene expression. We computationally and experimentally identify a major role of these physical factors, specifically chromatin packing scaling, in regulating phenotypic plasticity, determining responsiveness to external stressors by influencing both intercellular transcriptional malleability and heterogeneity. Applying CPMC model predictions to transcriptional data from cancer patients, we identify an inverse relationship between patient survival and phenotypic plasticity of tumor cells.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures

References
-
- Rathert P., Roth M., Neumann T., Muerdter F., Roe J.-S., Muhar M., Deswal S., Cerny-Reiterer S., Peter B., Jude J., Hoffmann T., Boryń Ł. M., Axelsson E., Schweifer N., Tontsch-Grunt U., Dow L. E., Gianni D., Pearson M., Valent P., Stark A., Kraut N., Vakoc C. R., Zuber J., Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature 525, 543–547 (2015). - PMC - PubMed
-
- Nguyen Q. H., Pervolarakis N., Blake K., Ma D., Davis R. T., James N., Phung A. T., Willey E., Kumar R., Jabart E., Driver I., Rock J., Goga A., Khan S. A., Lawson D. A., Werb Z., Kessenbrock K., Profiling human breast epithelial cells using single cell RNA sequencing identifies cell diversity. Nat. Commun. 9, 2028 (2018). - PMC - PubMed
-
- Haber A. L., Biton M., Rogel N., Herbst R. H., Shekhar K., Smillie C., Burgin G., Delorey T. M., Howitt M. R., Katz Y., Tirosh I., Beyaz S., Dionne D., Zhang M., Raychowdhury R., Garrett W. S., Rozenblatt-Rosen O., Shi H. N., Yilmaz O., Xavier R. J., Regev A., A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

