A meta-analysis of genome-wide association studies of multiple myeloma among men and women of African ancestry
- PMID: 31935283
- PMCID: PMC6960456
- DOI: 10.1182/bloodadvances.2019000491
A meta-analysis of genome-wide association studies of multiple myeloma among men and women of African ancestry
Abstract
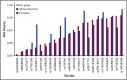
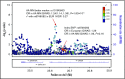
Persons of African ancestry (AA) have a twofold higher risk for multiple myeloma (MM) compared with persons of European ancestry (EA). Genome-wide association studies (GWASs) support a genetic contribution to MM etiology in individuals of EA. Little is known about genetic risk factors for MM in individuals of AA. We performed a meta-analysis of 2 GWASs of MM in 1813 cases and 8871 controls and conducted an admixture mapping scan to identify risk alleles. We fine-mapped the 23 known susceptibility loci to find markers that could better capture MM risk in individuals of AA and constructed a polygenic risk score (PRS) to assess the aggregated effect of known MM risk alleles. In GWAS meta-analysis, we identified 2 suggestive novel loci located at 9p24.3 and 9p13.1 at P < 1 × 10-6; however, no genome-wide significant association was noted. In admixture mapping, we observed a genome-wide significant inverse association between local AA at 2p24.1-23.1 and MM risk in AA individuals. Of the 23 known EA risk variants, 20 showed directional consistency, and 9 replicated at P < .05 in AA individuals. In 8 regions, we identified markers that better capture MM risk in persons with AA. AA individuals with a PRS in the top 10% had a 1.82-fold (95% confidence interval, 1.56-2.11) increased MM risk compared with those with average risk (25%-75%). The strongest functional association was between the risk allele for variant rs56219066 at 5q15 and lower ELL2 expression (P = 5.1 × 10-12). Our study shows that common genetic variation contributes to MM risk in individuals with AA.
Conflict of interest statement
Conflict-of-interest disclosure: C. A. Huff has acted as a consultant for Karyopharm Therapeutics, Sanofi, and miDiagnostics and is a member of the Safety Monitoring Board for Johnson and Johnson. T.G.M. has acted as a consultant for Roche and Juno Therapeutics and has received research funding from Amgen, Sanofi, and Seattle Genetics. J.M. is a member of the Speakers Bureau for Takeda Pharmacauticals and Celgene and owns stock in Celgene, Bristol-Myers Squibb, and bluebird bio. S.S. is a member of the Speakers Bureau for Takeda Pharmaceuticals, Celgene, and Janssen Pharmaceuticals and owns stock in Celgene, Bristol-Myers Squibb, and bluebird bio. S.L. has acted as a consultant for Janssen Pharmaceuticals, Takeda Pharmaceuticals, Celgene, Novartis, Bristol-Myers Squibb, Merck, and GlaxoSmithKline and has received research funding from Celgene, Takeda Pharmaceuticals, and Janssen Pharmaceuticals. K.C.A. is a consultant for Celgene, Jansen, Bristol-Myers Squibb and Sanofi, and a scientific founder of OncoPep and C4 Therapeutics. S.A. has acted as a consultant for Novartis, Amgen, and Takeda Pharmaceuticals, and has received research funding from Pharmacyclics. A.K.N. has acted as a consultant for Amgen, Novartis, Spectrum Pharmaceuticals, and Adaptive Biotechnologies. R.V. has received honoraria and research funding from Takeda Pharmaceuticals and Amgen and received honoraria from Celgene, Bristol-Myers Squibb, Janssen Pharmaceuticals, AbbVie, Jazz Pharmaceuticals, and Konypharma. J.A.Z. has acted as a consultant for Prothena and Janssen Pharmaceuticals; has acted as a consultant and received research funding from Bristol-Myers Squibb, Celgene, and Takeda Pharmaceuticals; and is a member of the Data Safety Monitoring Committee for Pharmacyclics. G.J.M. has acted as a consultant for Celgene, Takeda Phamaceuticals, and Bristol-Myers Squibb; has received research funding from Celgene; and has received honoraria from Celgene, Takeda Pharmaceuticals, and Bristol-Myers Squibb. R.Z.O. is a member of the Advisory Board for Amgen, Celgene, Forma Therapeutics, GlaxoSmithKline Biologicals, Ionis Pharmaceuticals, Janssen Biotech, Juno Therapeutics, Kite Pharma, Legend Biotech, Sanofi, Servier, and Takeda Pharmaceuticals; has acted as a consultant for Molecular Partners; and has received research funding from BioTheryX. The remaining authors declare no competing financial interests.
Figures

References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7-30. - PubMed
-
- Gebregziabher M, Bernstein L, Wang Y, Cozen W. Risk patterns of multiple myeloma in Los Angeles County, 1972-1999 (United States). Cancer Causes Control. 2006;17(7):931-938. - PubMed
-
- Grufferman S, Cohen HJ, Delzell ES, Morrison MC, Schold SC Jr, Moore JO. Familial aggregation of multiple myeloma and central nervous system diseases. J Am Geriatr Soc. 1989;37(4):303-309. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 CA191896/CA/NCI NIH HHS/United States
- R01 CA084979/CA/NCI NIH HHS/United States
- R01 CA134786/CA/NCI NIH HHS/United States
- P30 CA006973/CA/NCI NIH HHS/United States
- UG1 CA189974/CA/NCI NIH HHS/United States
- U58 DP000807/DP/NCCDPHP CDC HHS/United States
- P30 CA014089/CA/NCI NIH HHS/United States
- P50 CA100707/CA/NCI NIH HHS/United States
- UG1 CA233163/CA/NCI NIH HHS/United States
- P2C HD050924/HD/NICHD NIH HHS/United States
- U10 CA037429/CA/NCI NIH HHS/United States
- U01 CA182883/CA/NCI NIH HHS/United States
- R01 CA092447/CA/NCI NIH HHS/United States
- P50 CA171963/CA/NCI NIH HHS/United States
- UM1 CA182883/CA/NCI NIH HHS/United States
- U01 CA164973/CA/NCI NIH HHS/United States

