Treatment-Induced Tumor Dormancy through YAP-Mediated Transcriptional Reprogramming of the Apoptotic Pathway
- PMID: 31935369
- PMCID: PMC7146079
- DOI: 10.1016/j.ccell.2019.12.006
Treatment-Induced Tumor Dormancy through YAP-Mediated Transcriptional Reprogramming of the Apoptotic Pathway
Abstract
Eradicating tumor dormancy that develops following epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) treatment of EGFR-mutant non-small cell lung cancer, is an attractive therapeutic strategy but the mechanisms governing this process are poorly understood. Blockade of ERK1/2 reactivation following EGFR TKI treatment by combined EGFR/MEK inhibition uncovers cells that survive by entering a senescence-like dormant state characterized by high YAP/TEAD activity. YAP/TEAD engage the epithelial-to-mesenchymal transition transcription factor SLUG to directly repress pro-apoptotic BMF, limiting drug-induced apoptosis. Pharmacological co-inhibition of YAP and TEAD, or genetic deletion of YAP1, all deplete dormant cells by enhancing EGFR/MEK inhibition-induced apoptosis. Enhancing the initial efficacy of targeted therapies could ultimately lead to prolonged treatment responses in cancer patients.
Keywords: YAP; dormancy; drug resistance; drug tolerance; epidermal growth factor receptor; lung cancer; senescence.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures
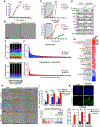

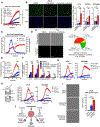
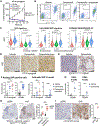


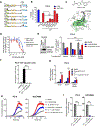

Comment in
-
Senesce to Survive: YAP-Mediated Dormancy Escapes EGFR/MEK Inhibition.Cancer Cell. 2020 Jan 13;37(1):1-2. doi: 10.1016/j.ccell.2019.12.008. Cancer Cell. 2020. PMID: 31951560
-
YAP Mediates EGFR Inhibitor-Induced Dormancy in EGFR-Mutant Cancer.Cancer Discov. 2020 Mar;10(3):OF2. doi: 10.1158/2159-8290.CD-RW2020-013. Epub 2020 Jan 24. Cancer Discov. 2020. PMID: 31980569
References
-
- Alexander WM, Ficarro SB, Adelmant G, and Marto JA (2017). multiplierz v2.0: A Python-based ecosystem for shared access and analysis of native mass spectrometry data. Proteomics 17, 15–16. - PubMed
-
- Beyer TA, Weiss A, Khomchuk Y, Huang K, Ogunjimi AA, Varelas X, and Wrana JL (2013). Switch enhancers interpret TGF-β and hippo signaling to control cell fate in human embryonic stem cells. Cell Rep. 5, 1611–1624. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

