Structure Modeling of the Norepinephrine Transporter
- PMID: 31936154
- PMCID: PMC7022499
- DOI: 10.3390/biom10010102
Structure Modeling of the Norepinephrine Transporter
Abstract
The norepinephrine transporter (NET) is one of the monoamine transporters. Its X-ray crystal structure has not been obtained yet. Inhibitors of human NET (hNET) play a major role in the treatment of many central and peripheral nervous system diseases. In this study, we focused on the spatial structure of a NET constructed by homology modeling on Drosophila melanogaster dopamine transporter templates. We further examined molecular construction of primary binding pocket (S1) together with secondary binding site (S2) and extracellular loop 4 (EL4). The next stage involved docking of transporter inhibitors: Reboxetine, duloxetine, desipramine, and other commonly used drugs. The procedure revealed the molecular orientation of residues and disclosed ones that are the most important for ligand binding: Phenylalanine F72, aspartic acid D75, tyrosine Y152, and phenylalanine F317. Aspartic acid D75 plays a key role in recognition of the basic amino group present in monoamine transporter inhibitors and substrates. The study also presents a comparison of hNET models with other related proteins, which could provide new insights into their interaction with therapeutics and aid future development of novel bioactive compounds.
Keywords: homology modeling; ligand docking; norepinephrine transporter; reuptake inhibitors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures













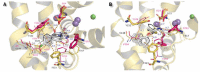
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

