Comparative Genomics Analysis of Lactobacillus ruminis from Different Niches
- PMID: 31936280
- PMCID: PMC7016997
- DOI: 10.3390/genes11010070
Comparative Genomics Analysis of Lactobacillus ruminis from Different Niches
Abstract
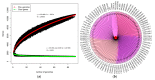
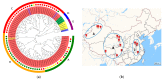
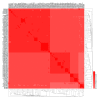
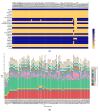
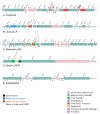
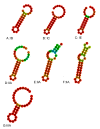
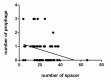
Lactobacillus ruminis is a commensal motile lactic acid bacterium living in the intestinal tract of humans and animals. Although a few genomes of L. ruminis were published, most of them were animal derived. To explore the genetic diversity and potential niche-specific adaptation changes of L. ruminis, in the current work, draft genomes of 81 L. ruminis strains isolated from human, bovine, piglet, and other animals were sequenced, and comparative genomic analysis was performed. The genome size and GC content of L. ruminis on average were 2.16 Mb and 43.65%, respectively. Both the origin and the sampling distance of these strains had a great influence on the phylogenetic relationship. For carbohydrate utilization, the human-derived L. ruminis strains had a higher consistency in the utilization of carbon source compared to the animal-derived strains. L. ruminis mainly increased the competitiveness of niches by producing class II bacteriocins. The type of clustered regularly interspaced short palindromic repeats /CRISPR-associated (CRISPR/Cas) system presented in L. ruminis was mainly subtype IIA. The diversity of CRISPR/Cas locus depended on the high denaturation of spacer number and sequence, although cas1 protein was relatively conservative. The genetic differences in those newly sequenced L. ruminis strains highlighted the gene gains and losses attributed to niche adaptations.
Keywords: CRISPR/Cas; Lactobacillus ruminis; bacteriocins; carbohydrate utilization; phylogenetic relationship; prophage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Lactobacillus ruminis strains cluster according to their mammalian gut source.BMC Microbiol. 2015 Apr 1;15:80. doi: 10.1186/s12866-015-0403-y. BMC Microbiol. 2015. PMID: 25879663 Free PMC article.
-
Genome sequences and comparative genomics of two Lactobacillus ruminis strains from the bovine and human intestinal tracts.Microb Cell Fact. 2011 Aug 30;10 Suppl 1(Suppl 1):S13. doi: 10.1186/1475-2859-10-S1-S13. Epub 2011 Aug 30. Microb Cell Fact. 2011. PMID: 21995554 Free PMC article.
-
Comparative Genomics Analysis of Lactobacillus mucosae from Different Niches.Genes (Basel). 2020 Jan 14;11(1):95. doi: 10.3390/genes11010095. Genes (Basel). 2020. PMID: 31947593 Free PMC article.
-
[Why so rare if so essentiel: the determinants of the sparse distribution of CRISPR-Cas systems in bacterial genomes].Biol Aujourdhui. 2017;211(4):255-264. doi: 10.1051/jbio/2018005. Epub 2018 Jun 29. Biol Aujourdhui. 2017. PMID: 29956652 Review. French.
-
Harnessing CRISPR-Cas systems for bacterial genome editing.Trends Microbiol. 2015 Apr;23(4):225-32. doi: 10.1016/j.tim.2015.01.008. Epub 2015 Feb 17. Trends Microbiol. 2015. PMID: 25698413 Review.
Cited by
-
Lactobacillus ruminis Alleviates DSS-Induced Colitis by Inflammatory Cytokines and Gut Microbiota Modulation.Foods. 2021 Jun 11;10(6):1349. doi: 10.3390/foods10061349. Foods. 2021. PMID: 34208038 Free PMC article.
-
Integrated Phenotypic-Genotypic Analysis of Latilactobacillus sakei from Different Niches.Foods. 2021 Jul 25;10(8):1717. doi: 10.3390/foods10081717. Foods. 2021. PMID: 34441495 Free PMC article.
-
Assessment of beneficial effects and identification of host adaptation-associated genes of Ligilactobacillus salivarius isolated from badgers.BMC Genomics. 2023 Sep 7;24(1):530. doi: 10.1186/s12864-023-09623-8. BMC Genomics. 2023. PMID: 37679681 Free PMC article.
-
Identification and Characterization of the CRISPR/Cas System in Staphylococcus aureus Strains From Diverse Sources.Front Microbiol. 2021 Jun 2;12:656996. doi: 10.3389/fmicb.2021.656996. eCollection 2021. Front Microbiol. 2021. PMID: 34149645 Free PMC article.
-
Determination of Technological Properties and CRISPR Profiles of Streptococcus thermophilus Isolates Obtained from Local Yogurt Samples.Microorganisms. 2024 Nov 25;12(12):2428. doi: 10.3390/microorganisms12122428. Microorganisms. 2024. PMID: 39770631 Free PMC article.
References
-
- Hammes W.P., Hertel C. Lactobacillus. In: Whitman W.B., editor. Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2015. pp. 1–76. - DOI
-
- Lerche M., Reuter G. A contribution to the method of isolation and differentiation of aerobic “lactobacilli” (Genus “Lactobacillus beijerinck”) Zent. Bakteriol. Int. J. Med Microbiol. 1960;179:354–370. - PubMed
-
- O’Donnell M.M., Forde B.M., Neville B., Ross P.R., O’Toole P.W. Carbohydrate catabolic flexibility in the mammalian intestinal commensal Lactobacillus ruminis revealed by fermentation studies aligned to genome annotations. Microb. Cell Fact. 2011;10(Suppl. S1):S12. doi: 10.1186/1475-2859-10-S1-S12. - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

