STIM Protein-NMDA2 Receptor Interaction Decreases NMDA-Dependent Calcium Levels in Cortical Neurons
- PMID: 31936514
- PMCID: PMC7017226
- DOI: 10.3390/cells9010160
STIM Protein-NMDA2 Receptor Interaction Decreases NMDA-Dependent Calcium Levels in Cortical Neurons
Abstract
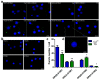
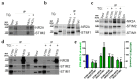
Neuronal Store-Operated Ca2+ Entry (nSOCE) plays an essential role in refilling endoplasmic reticulum Ca2+ stores and is critical for Ca2+-dependent neuronal processes. SOCE sensors, STIM1 and STIM2, can activate Orai, TRP channels and AMPA receptors, and inhibit voltage-gated channels in the plasma membrane. However, the link between STIM, SOCE, and NMDA receptors, another key cellular entry point for Ca2+ contributing to synaptic plasticity and excitotoxicity, remains unclear. Using Ca2+ imaging, we demonstrated that thapsigargin-induced nSOCE was inhibited in rat cortical neurons following NMDAR inhibitors. Blocking nSOCE by its inhibitor SKF96365 enhanced NMDA-driven [Ca2+]i. Modulating STIM protein level through overexpression or shRNA inhibited or activated NMDA-evoked [Ca2+]i, respectively. Using proximity ligation assays, immunofluorescence, and co-immunoprecipitation methods, we discovered that thapsigargin-dependent effects required interactions between STIMs and the NMDAR2 subunits. Since STIMs modulate NMDAR-mediated Ca2+ levels, we propose targeting this mechanism as a novel therapeutic strategy against neuropathological conditions that feature NMDA-induced Ca2+ overload as a diagnostic criterion.
Keywords: Ca2+ homeostasis; NMDA receptor; STIM proteins; endoplasmic reticulum (ER); neuronal store-operated calcium entry (nSOCE); neurons; organellar Ca2+; plasma membrane (PM).
Conflict of interest statement
The authors declare no conflict of interest and that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

