Synthetic DNA Vaccines Adjuvanted with pIL-33 Drive Liver-Localized T Cells and Provide Protection from Plasmodium Challenge in a Mouse Model
- PMID: 31936739
- PMCID: PMC7157753
- DOI: 10.3390/vaccines8010021
Synthetic DNA Vaccines Adjuvanted with pIL-33 Drive Liver-Localized T Cells and Provide Protection from Plasmodium Challenge in a Mouse Model
Abstract
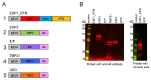
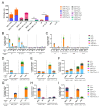
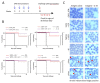
The need for a malaria vaccine is indisputable. A single vaccine for Plasmodium pre-erythrocytic stages targeting the major sporozoite antigen circumsporozoite protein (CSP) has had partial success. Additionally, CD8+ T cells targeting liver-stage (LS) antigens induced by live attenuated sporozoite vaccines were associated with protection in human challenge experiments. To further evaluate protection mediated by LS antigens, we focused on exported pre-erythrocytic proteins (exported protein 1 (EXP1), profilin (PFN), exported protein 2 (EXP2), inhibitor of cysteine proteases (ICP), transmembrane protein 21 (TMP21), and upregulated in infective sporozoites-3 (UIS3)) expressed in all Plasmodium species and designed optimized, synthetic DNA (synDNA) immunogens. SynDNA antigen cocktails were tested with and without the molecular adjuvant plasmid IL-33. Immunized animals developed robust T cell responses including induction of antigen-specific liver-localized CD8+ T cells, which were enhanced by the co-delivery of plasmid IL-33. In total, 100% of mice in adjuvanted groups and 71%-88% in non-adjuvanted groups were protected from blood-stage disease following Plasmodium yoelii sporozoite challenge. This study supports the potential of synDNA LS antigens as vaccine components for malaria parasite infection.
Keywords: DNA vaccines; Plasmodium; exported proteins; liver stage; malaria.
Conflict of interest statement
The authors declare the following conflicts of interest: D.B.W. reports receiving commercial research funding from Inovio, Geneos, and GeneOne; has received speakers bureau honoraria from AstraZeneca, Roche and Merck; has ownership interest in Inovio; and is a consultant/board member for Inovio. L.H. is an employee of Inovio Pharmaceuticals. No potential conflicts of interests were disclosed by the other authors.
Figures



Similar articles
-
An Attenuated HSV-1-Derived Malaria Vaccine Expressing Liver-Stage Exported Proteins Induces Sterilizing Protection against Infectious Sporozoite Challenge.Vaccines (Basel). 2022 Feb 16;10(2):300. doi: 10.3390/vaccines10020300. Vaccines (Basel). 2022. PMID: 35214758 Free PMC article.
-
Defining rules of CD8(+) T cell expansion against pre-erythrocytic Plasmodium antigens in sporozoite-immunized mice.Malar J. 2016 Apr 26;15:238. doi: 10.1186/s12936-016-1295-5. Malar J. 2016. PMID: 27113469 Free PMC article.
-
Protracted sterile protection with Plasmodium yoelii pre-erythrocytic genetically attenuated parasite malaria vaccines is independent of significant liver-stage persistence and is mediated by CD8+ T cells.J Infect Dis. 2007 Aug 15;196(4):608-16. doi: 10.1086/519742. Epub 2007 Jul 9. J Infect Dis. 2007. PMID: 17624848
-
T cell responses to pre-erythrocytic stages of malaria: role in protection and vaccine development against pre-erythrocytic stages.Annu Rev Immunol. 1993;11:687-727. doi: 10.1146/annurev.iy.11.040193.003351. Annu Rev Immunol. 1993. PMID: 8476576 Review.
-
Pre-erythrocytic malaria vaccine: mechanisms of protective immunity and human vaccine trials.Parassitologia. 1999 Sep;41(1-3):397-402. Parassitologia. 1999. PMID: 10697892 Review.
Cited by
-
An Attenuated HSV-1-Derived Malaria Vaccine Expressing Liver-Stage Exported Proteins Induces Sterilizing Protection against Infectious Sporozoite Challenge.Vaccines (Basel). 2022 Feb 16;10(2):300. doi: 10.3390/vaccines10020300. Vaccines (Basel). 2022. PMID: 35214758 Free PMC article.
-
Harnessing Recent Advances in Synthetic DNA and Electroporation Technologies for Rapid Vaccine Development Against COVID-19 and Other Emerging Infectious Diseases.Front Med Technol. 2020 Oct 21;2:571030. doi: 10.3389/fmedt.2020.571030. eCollection 2020. Front Med Technol. 2020. PMID: 35047878 Free PMC article. Review.
-
Subcutaneous Immunization with Unaltered Axenic Malaria Parasite Liver Stages Induces Sterile Protection against Infectious Sporozoite Challenge.Vaccines (Basel). 2022 Nov 8;10(11):1884. doi: 10.3390/vaccines10111884. Vaccines (Basel). 2022. PMID: 36366392 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

