CRISPR- cas3 of Salmonella Upregulates Bacterial Biofilm Formation and Virulence to Host Cells by Targeting Quorum-Sensing Systems
- PMID: 31936769
- PMCID: PMC7168661
- DOI: 10.3390/pathogens9010053
CRISPR- cas3 of Salmonella Upregulates Bacterial Biofilm Formation and Virulence to Host Cells by Targeting Quorum-Sensing Systems
Abstract
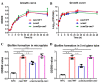
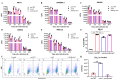
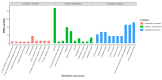
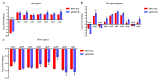
Salmonella is recognized as one of the most common microbial pathogens worldwide. The bacterium contains the clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated (Cas) systems, providing adaptive immunity against invading foreign nucleic acids. Previous studies suggested that certain bacteria employ the Cas proteins of CRISPR-Cas systems to target their own genes, which also alters the virulence during invasion of mammals. However, whether CRISPR-Cas systems in Salmonella have similar functions during bacterial invasion of host cells remains unknown. Here, we systematically analyzed the genes that are regulated by Cas3 in a type I-E CRISPR-Cas system and the virulence changes due to the deletion of cas3 in Salmonella enterica serovar Enteritidis. Compared to the cas3 gene wild-type (cas3 WT) Salmonella strain, cas3 deletion upregulated the lsrFGBE genes in lsr (luxS regulated) operon related to quorum sensing (QS) and downregulated biofilm-forming-related genes and Salmonella pathogenicity island 1 (SPI-1) genes related to the type three secretion system (T3SS). Consistently, the biofilm formation ability was downregulated in the cas3 deletion mutant (Δcas3). The bacterial invasive and intracellular capacity of Δcas3 to host cells was also reduced, thereby increasing the survival of infected host cells and live chickens. By the transcriptome-wide screen (RNA-Seq), we found that the cas3 gene impacts a series of genes related to QS, the flagellum, and SPI-1-T3SS system, thereby altering the virulence phenotypes. As QS SPI-1-T3SS and CRISPR-Cas systems are widely distributed in the bacteria kingdom, our findings extend our understanding of virulence regulation and pathogenicity in mammalian hosts for Salmonella and potentially other bacteria.
Keywords: RNA-Seq; SPI-1-T3SS; Salmonella virulence; biofilm formation; cas3; quorum sensing; type I-E CRISPR-Cas system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Hohmann E.L. Nontyphoidal salmonellosis. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2001;32:263–269. - PubMed
-
- Chen Z., Zhang K., Yin H., Li Q., Wang L., Liu Z. Detection of Salmonella and several common Salmonella serotypes in food by loop-mediated isothermal amplification method. Food Sci. Hum. Wellness. 2015;4:75–79. doi: 10.1016/j.fshw.2015.05.001. - DOI
Grants and funding
- P20GM103442/Foundation for the National Institutes of Health
- 2662017JC034/Fundamental Research Funds for the Central Universities
- AI101973-01/Foundation for the National Institutes of Health
- AI109317-01A1/Foundation for the National Institutes of Health
- 2017020201010228/Applied Basic Research Programs of Wuhan
- CARS-35/Earmarked Fund for China Agriculture Research System
- 2017YFC1600100/National Basic Research Program of China (973 Program)
- P20GM113123/Foundation for the National Institutes of Health
- P20 GM103442/GM/NIGMS NIH HHS/United States
- AI097532-01A1/Foundation for the National Institutes of Health
- P20 GM113123/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

