The U-Rich Untranslated Region of the Hepatitis E Virus Induces Differential Type I and Type III Interferon Responses in a Host Cell-Dependent Manner
- PMID: 31937650
- PMCID: PMC6960293
- DOI: 10.1128/mBio.03103-19
The U-Rich Untranslated Region of the Hepatitis E Virus Induces Differential Type I and Type III Interferon Responses in a Host Cell-Dependent Manner
Abstract
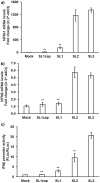
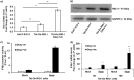
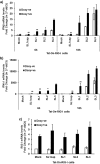
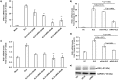
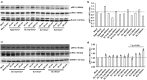
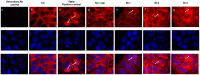
Hepatitis E virus (HEV), a single-strand positive-sense RNA virus, is an understudied but important human pathogen. The virus can establish infection at a number of host tissues, including the small intestine and liver, causing acute and chronic hepatitis E as well as certain neurological disorders. The retinoic acid-inducible gene I (RIG-I) pathway is essential to induce the interferon (IFN) response during HEV infection. However, the pathogen-associated motif patterns (PAMPs) in the HEV genome that are recognized by RIG-I remain unknown. In this study, we first identified that HEV RNA PAMPs derived from the 3' untranslated region (UTR) of the HEV genome induced higher levels of IFN mRNA, interferon regulatory factor-3 (IRF3) phosphorylation, and nuclear translocation than the 5' UTR of HEV. We revealed that the U-rich region in the 3' UTR of the HEV genome acts as a potent RIG-I PAMP, while the presence of poly(A) tail in the 3' UTR further increases the potency. We further demonstrated that HEV UTR PAMPs induce differential type I and type III IFN responses in a cell type-dependent fashion. Predominant type III IFN response was observed in the liver tissues of pigs experimentally infected with HEV as well as in HEV RNA PAMP-induced human hepatocytes in vitro In contrast, HEV RNA PAMPs induced a predominant type I IFN response in swine enterocytes. Taken together, the results from this study indicated that the IFN response during HEV infection depends both on viral RNA motifs and host target cell types. The results have important implications in understanding the mechanism of HEV pathogenesis.IMPORTANCE Hepatitis E virus (HEV) is an important human pathogen causing both acute and chronic viral hepatitis E infection. Currently, the mechanisms of HEV replication and pathogenesis remain poorly understood. The innate immune response acts as the first line of defense during viral infection. The retinoic acid-inducible gene I (RIG-I)-mediated interferon (IFN) response has been implicated in establishing antiviral response during HEV infection, although the HEV RNA motifs that are recognized by RIG-I are unknown. This study identified that the U-rich region in the 3' untranslated region (UTR) of the HEV genome acts as a potent RIG-I agonist compared to the HEV 5' UTR. We further revealed that the HEV RNA pathogen-associated motif patterns (PAMPs) induced a differential IFN response in a cell type-dependent manner: a predominantly type III IFN response in hepatocytes, and a predominantly type I IFN response in enterocytes. These data demonstrate the complexity by which both host and viral factors influence the IFN response during HEV infection.
Keywords: U-rich region RNA PAMPs; hepatitis E virus (HEV); retinoic acid-inducible gene I (RIG-I); type I interferon (IFN); type-III IFN.
Copyright © 2020 Sooryanarain et al.
Figures
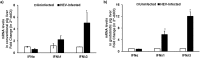


Similar articles
-
Identification of the RNA Pseudoknot within the 3' End of the Porcine Reproductive and Respiratory Syndrome Virus Genome as a Pathogen-Associated Molecular Pattern To Activate Antiviral Signaling via RIG-I and Toll-Like Receptor 3.J Virol. 2018 May 29;92(12):e00097-18. doi: 10.1128/JVI.00097-18. Print 2018 Jun 15. J Virol. 2018. PMID: 29618647 Free PMC article.
-
RIG-I is a key antiviral interferon-stimulated gene against hepatitis E virus regardless of interferon production.Hepatology. 2017 Jun;65(6):1823-1839. doi: 10.1002/hep.29105. Epub 2017 May 3. Hepatology. 2017. PMID: 28195391
-
Pathogen-Associated Molecular Pattern Recognition of Hepatitis C Virus Transmitted/Founder Variants by RIG-I Is Dependent on U-Core Length.J Virol. 2015 Nov;89(21):11056-68. doi: 10.1128/JVI.01964-15. Epub 2015 Aug 26. J Virol. 2015. PMID: 26311867 Free PMC article.
-
Hepatocyte Intrinsic Innate Antiviral Immunity against Hepatitis Delta Virus Infection: The Voices of Bona Fide Human Hepatocytes.Viruses. 2024 May 8;16(5):740. doi: 10.3390/v16050740. Viruses. 2024. PMID: 38793622 Free PMC article. Review.
-
Host Innate Immunity against Hepatitis E Virus and Viral Evasion Mechanisms.J Microbiol Biotechnol. 2017 Oct 28;27(10):1727-1735. doi: 10.4014/jmb.1708.08045. J Microbiol Biotechnol. 2017. PMID: 29017236 Review.
Cited by
-
Immunobiology and Host Response to HEV.Adv Exp Med Biol. 2023;1417:93-118. doi: 10.1007/978-981-99-1304-6_7. Adv Exp Med Biol. 2023. PMID: 37223861
-
Modulation of SOCS3 Levels via STAT3 and Estrogen-ERαp66 Signaling during Hepatitis E Virus Replication in Hepatocellular Carcinoma Cells.J Virol. 2022 Oct 12;96(19):e0100822. doi: 10.1128/jvi.01008-22. Epub 2022 Sep 14. J Virol. 2022. PMID: 36102649 Free PMC article.
-
Emerging RNA-centric technologies to probe RNA-protein interactions: importance in decoding the life cycle of positive sense single strand RNA viruses and antiviral discovery.Front Cell Infect Microbiol. 2025 May 21;15:1580337. doi: 10.3389/fcimb.2025.1580337. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40584171 Free PMC article. Review.
-
Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors.Cell Mol Immunol. 2021 Mar;18(3):539-555. doi: 10.1038/s41423-020-00602-7. Epub 2021 Jan 18. Cell Mol Immunol. 2021. PMID: 33462384 Free PMC article. Review.
-
The Re-Emergence of Hepatitis E Virus in Europe and Vaccine Development.Viruses. 2023 Jul 16;15(7):1558. doi: 10.3390/v15071558. Viruses. 2023. PMID: 37515244 Free PMC article. Review.
References
-
- Smith DB, Simmonds P, members of the International Committee on the Taxonomy of Viruses Hepeviridae Study Group, Jameel S, Emerson SU, Harrison TJ, Meng XJ, Okamoto H, Van der Poel WH, Purdy MA. 2015. Consensus proposals for classification of the family Hepeviridae. J Gen Virol 96:1191–1192. doi:10.1099/vir.0.000115. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

