Bioinformatic analysis of differential expression and core GENEs in breast cancer
- PMID: 31938209
- PMCID: PMC6958129
Bioinformatic analysis of differential expression and core GENEs in breast cancer
Abstract
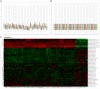
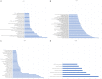
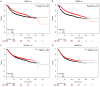
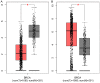
Breast cancer (BRCA) is one of the most common malignancies in women. The gene expression profile of GSE103512 from the GEO database was downloaded in order to find key genes involved in the occurrence and development of BRCA. 75 samples, including 65 cancer and 10 normal samples, were included in this analysis. Differentially expressed genes (DEGs) between BRCA patients and health people were chosen using R tool. We next performed gene ontology (GO) analysis and Kyoto Encyclopedia of Gene and Genome (KEGG) pathway analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID). Moreover, Cytoscape with Search Tool for the Retrieval of Interacting Genes (STRING) was utilized to visualize protein-protein interaction (PPI) of these DEGs. The related genes and medicines specific to hub genes were predicted by CBioportal. We screened a total of 357 DEGs including 77 up-regulated and 280 down-regulated. A series of BRCA related GO terms and pathways were identified by analysis of these DEGs. Insulin-like growth factor 1 (IGF1); epidermal growth factor receptor (EGFR); v-jun avian sarcoma virus 17 oncogene homolog (JUN) and Estrogen Receptor 1 (ESR1) of the DEGs were screened by construction of the PPI network and the degree of connectivity. IGF1 and ESR1 were finally selected as potential hub genes and treatment targets of BRCA. In conclusion, this bioinformatics analysis demonstrated that DEGs and hub genes, such as IGF1, might regulate the development of gastric cancer. These DEGs could be used as new biomarkers for diagnosis and to guide the combination medicine of BRCA.
Keywords: Breast cancer; bioinformatics analysis; biomarker; differential expression genes; therapeutic.
IJCEP Copyright © 2018.
Conflict of interest statement
None.
Figures


References
-
- DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64:52–62. - PubMed
-
- Donepudi MS, Kondapalli K, Amos SJ, Venkanteshan P. Breast cancer statistics and markers. J Cancer Res Ther. 2014;10:506–511. - PubMed
-
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. - PubMed
-
- DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66:31–42. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
