Detection of bladder cancer using urinary cell-free DNA and cellular DNA
- PMID: 31938901
- PMCID: PMC6960275
- DOI: 10.1186/s40169-020-0257-2
Detection of bladder cancer using urinary cell-free DNA and cellular DNA
Abstract
Background: The present study sought to identify a panel of DNA markers for noninvasive diagnosis using cell-free DNA (cfDNA) from urine supernatant or cellular DNA from urine sediments of hematuria patients. A panel of 48 bladder cancer-specific genes was selected. A next-generation sequencing-based assay with a cfDNA barcode-enabled single-molecule test was employed. Mutation profiles of blood, urine, and tumor sample from 16 bladder cancer patients were compared. Next, urinary cellular DNA and cfDNA were prospectively collected from 125 patients (92 bladder cancer cases and 33 controls) and analyzed using the 48-gene panel. The individual gene markers and combinations of markers were validated according to the pathology results. The mean areas under the receiver operating characteristic (ROC) curves (AUCs) obtained with the various modeling approaches were calculated and compared.
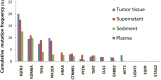
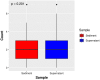
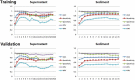
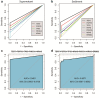
Results: This pilot study of 16 bladder cancer patients demonstrated that gene mutations in urine supernatant and sediments had better concordance with cancer tissue as compared with plasma. Logistic analyses suggested two powerful combinations of genes for genetic diagnostic modeling: five genes for urine supernatant (TERT, FGFR3, TP53, PIK3CA, and KRAS) and seven genes for urine sediments (TERT, FGFR3, TP53, HRAS, PIK3CA, KRAS, and ERBB2). The accuracy of the five-gene panel and the seven-gene panel in the validation cohort yielded AUCs of 0.94 [95% confidence interval (CI) 0.91-0.97] and 0.91 (95% CI 0.86-0.96), respectively. With the addition of age and gender, the diagnostic power of the urine supernatant five-gene model and the urine sediment seven-gene model improved as the revised AUCs were 0.9656 (95% CI 0.9368-0.9944) and 0.9587 (95% CI 0.9291-0.9883).
Conclusions: cfDNA from urine bears great diagnostic potential. A five-gene panel for urine supernatant and a seven-gene panel for urine sediments are promising options for identifying bladder cancer in hematuria patients.
Keywords: Bladder cancer; Cell-free DNA; Hematuria; Mutation; Next-generation sequencing.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
