Signature Fragment Ions of Biotinylated Peptides
- PMID: 31939678
- PMCID: PMC7199424
- DOI: 10.1021/jasms.9b00024
Signature Fragment Ions of Biotinylated Peptides
Abstract
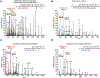
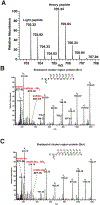
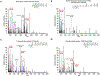
The use of biotin or biotin-containing reagents is an essential component of many protein purification and labeling technologies. Owing to its small size and high affinity to the avidin family of proteins, biotin is a versatile molecular handle that permits both enrichment and purity that is not easily achieved by other reagents. Traditionally, the use of biotinylation to enrich for proteins has not required the detection of the site of biotinylation. However, newer technologies for discovery of protein-protein interactions, such as APEX and BioID, as well as some of the click chemistry-based labeling approaches have underscored the importance of determining the exact residue that is modified by biotin. Anti-biotin antibody-based enrichment of biotinylated peptides (e.g., BioSITe) coupled to LC-MS/MS permit large-scale detection and localization of sites of biotinylation. As with any chemical modification of peptides, understanding the fragmentation patterns that result from biotin modification is essential to improving its detection by LC-MS/MS. Tandem mass spectra of biotinylated peptides has not yet been studied systematically. Here, we describe the various signature fragment ions generated with collision-induced dissociation of biotinylated peptides. We focused on biotin adducts attached to peptides generated by BioID and APEX experiments, including biotin, isotopically heavy biotin, and biotin-XX-phenol, a nonpermeable variant of biotin-phenol. We also highlight how the detection of biotinylated peptides in high-throughput studies poses certain computational challenges for accurate quantitation which need to be addressed. Our findings about signature fragment ions of biotinylated peptides should be helpful in the confirmation of biotinylation sites.
Keywords: marker ions; mass spectrometry; post-translational modifications; protein−protein interactions.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Chaiet L; Wolf FJ The Properties of Streptavidin, a Biotin-Binding Protein Produced by Streptomycetes. Arch. Biochem. Biophys 1964, 106, 1–5. - PubMed
-
- Cronan JE Jr. Biotination of proteins in vivo. A post-translational modification to label, purify, and study proteins. J. Biol. Chem 1990, 265 (18), 10327–33. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

