FOXD3 Regulates VISTA Expression in Melanoma
- PMID: 31940493
- PMCID: PMC6995351
- DOI: 10.1016/j.celrep.2019.12.036
FOXD3 Regulates VISTA Expression in Melanoma
Abstract
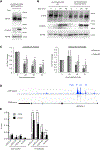
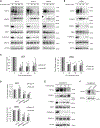
Immune checkpoint inhibitors have improved patient survival in melanoma, but the innate resistance of many patients necessitates the investigation of alternative immune targets. Many immune checkpoint proteins lack proper characterization, including V-domain Ig suppressor of T cell activation (VISTA). VISTA expression on immune cells can suppress T cell activity; however, few studies have investigated its expression and regulation in cancer cells. In this study, we observe that VISTA is expressed in melanoma patient samples and cell lines. Tumor cell-specific expression of VISTA promotes tumor onset in vivo, associated with increased intratumoral T regulatory cells, and enhanced PDL-1 expression on tumor-infiltrating macrophages. VISTA transcript levels are regulated by the stemness factor Forkhead box D3 (FOXD3). BRAF inhibition upregulates FOXD3 and reduces VISTA expression. Overall, this study demonstrates melanoma cell expression of VISTA and its regulation by FOXD3, contributing to the rationale for therapeutic strategies that combine targeted inhibitors with immune checkpoint blockade.
Keywords: DD1α; Dies1; FOXD3; PD-1H; VISTA; VSIR; immune checkpoint.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures
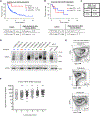

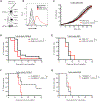
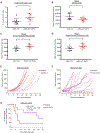
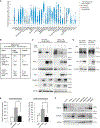
References
-
- Broad Institute TCGA Genome Data Analysis Center (2016). Broad GDAC Firehose 2016_01_28 run (Broad Institute of MIT and Harvard; ). 10.7908/C11G0KM9. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

