Subchromosome-Scale Nuclear and Complete Mitochondrial Genome Characteristics of Morchella crassipes
- PMID: 31940908
- PMCID: PMC7014384
- DOI: 10.3390/ijms21020483
Subchromosome-Scale Nuclear and Complete Mitochondrial Genome Characteristics of Morchella crassipes
Abstract
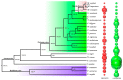
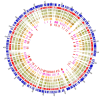
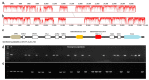
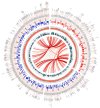
Morchella crassipes (Vent.) Pers., a typical yellow morel species with high economic value, is mainly distributed in the low altitude plains of Eurasia. However, rare research has been performed on its genomics and polarity, thus limiting its research and development. Here, we reported a fine physical map of the nuclear genome at the subchromosomal-scale and the complete mitochondrial genome of M. crassipes. The complete size of the nuclear genome was 56.7 Mb, and 23 scaffolds were assembled, with eight of them being complete chromosomes. A total of 11,565 encoding proteins were predicted. The divergence time analysis showed that M. crassipes representing yellow morels differentiated with black morels at ~33.98 Mya (million years), with 150 gene families contracted and expanded in M. crassipes versus the two black morels (M. snyderi and M. importuna). Furthermore, 409 CAZYme genes were annotated in M. crassipes, containing almost all plant cell wall degrading enzymes compared with the mycorrhizal fungi (truffles). Genomic annotation of mating type loci and amplification of the mating genes in the monospore population was conducted, the results indicated that M. crassipes is a heterothallic fungus. Additionally, a complete circular mitochondrial genome of M. crassipes was assembled, the size reached as large as 531,195 bp. It can be observed that the strikingly large size was the biggest up till now, coupled with 14 core conserved mitochondrial protein-coding genes, two rRNAs, 31 tRNAs, 51 introns, and 412 ncORFs. The total length of intron sequences accounted for 53.67% of the mitochondrial genome, with 19 introns having a length over 5 kb. Particularly, 221 of 412 ncORFs were distributed within 51 introns, and the total length of the ncORFs sequence accounted for 40.83% of the mitochondrial genome, and 297 ncORFs had expression activity in the mycelium stage, suggesting their potential functions in M. crassipes. Meanwhile, there was a high degree of repetition (51.31%) in the mitochondria of M. crassipes. Thus, the large number of introns, ncORFs and internal repeat sequences may contribute jointly to the largest fungal mitochondrial genome to date. The fine physical maps of nuclear genome and mitochondrial genome obtained in this study will open a new door for better understanding of the mysterious species of M. crassipes.
Keywords: Morchella crassipes; heterothallism; mating type; mitochondrial ORF; mitochondrial genome; nuclear genome.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
Similar articles
-
Opposite Polarity Monospore Genome De Novo Sequencing and Comparative Analysis Reveal the Possible Heterothallic Life Cycle of Morchella importuna.Int J Mol Sci. 2018 Aug 25;19(9):2525. doi: 10.3390/ijms19092525. Int J Mol Sci. 2018. PMID: 30149649 Free PMC article.
-
The mitochondrial genome of Morchella importuna (272.2 kb) is the largest among fungi and contains numerous introns, mitochondrial non-conserved open reading frames and repetitive sequences.Int J Biol Macromol. 2020 Jan 15;143:373-381. doi: 10.1016/j.ijbiomac.2019.12.056. Epub 2019 Dec 9. Int J Biol Macromol. 2020. PMID: 31830457
-
The whole-genome sequence analysis of Morchella sextelata.Sci Rep. 2019 Oct 25;9(1):15376. doi: 10.1038/s41598-019-51831-4. Sci Rep. 2019. PMID: 31653932 Free PMC article.
-
Mating Systems in True Morels (Morchella).Microbiol Mol Biol Rev. 2021 Aug 18;85(3):e0022020. doi: 10.1128/MMBR.00220-20. Epub 2021 Jul 28. Microbiol Mol Biol Rev. 2021. PMID: 34319143 Free PMC article. Review.
-
Artificial cultivation of true morels: current state, issues and perspectives.Crit Rev Biotechnol. 2018 Mar;38(2):259-271. doi: 10.1080/07388551.2017.1333082. Epub 2017 Jun 6. Crit Rev Biotechnol. 2018. PMID: 28585444 Review.
Cited by
-
Construction of nucleus-directed fluorescent reporter systems and its application to verification of heterokaryon formation in Morchella importuna.Front Microbiol. 2022 Nov 21;13:1051013. doi: 10.3389/fmicb.2022.1051013. eCollection 2022. Front Microbiol. 2022. PMID: 36478869 Free PMC article.
-
Genome assembly of M. spongiola and comparative genomics of the genus Morchella provide initial insights into taxonomy and adaptive evolution.BMC Genomics. 2024 May 27;25(1):518. doi: 10.1186/s12864-024-10418-8. BMC Genomics. 2024. Retraction in: BMC Genomics. 2025 May 7;26(1):449. doi: 10.1186/s12864-025-11667-x. PMID: 38802743 Free PMC article. Retracted.
-
Comparative mitochondrial genome analyses reveal conserved gene arrangement but massive expansion/contraction in two closely related Exserohilum pathogens.Comput Struct Biotechnol J. 2022 Mar 21;20:1456-1469. doi: 10.1016/j.csbj.2022.03.016. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35386100 Free PMC article.
-
Global Characterization of Fungal Mitogenomes: New Insights on Genomic Diversity and Dynamism of Coding Genes and Accessory Elements.Front Microbiol. 2021 Dec 1;12:787283. doi: 10.3389/fmicb.2021.787283. eCollection 2021. Front Microbiol. 2021. PMID: 34925295 Free PMC article.
-
Epidemic Identification of Fungal Diseases in Morchella Cultivation across China.J Fungi (Basel). 2022 Oct 20;8(10):1107. doi: 10.3390/jof8101107. J Fungi (Basel). 2022. PMID: 36294672 Free PMC article.
References
-
- Liu W., Zhang Y., He P. Morel Biology and Cultivation. Jilin science and Technology Press; Changchun, China: 2017.
-
- Pilz D., Rebecca M.L., Susan A., Luis V.R., Shannon B., Tricia W., Parks C.G., Erika M.F., Blaze B. Ecology and Management of Morels Harvested from the Forests of Western North America. United States Department of Agriculture; Washington, DC, USA: 2007.
-
- O’Donnell K., Rooney A.P., Mills G.L., Kuo M., Weber N.S., Rehner S.A. Phylogeny and historical biogeography of true morels (Morchella) reveals an early Cretaceous origin and high continental endemism and provincialism in the Holarctic. Fungal Genet. Biol. 2011;48:252–265. doi: 10.1016/j.fgb.2010.09.006. - DOI - PubMed
-
- Kuo M. Morels. University of Michigan Press; Ann Arbor, MI, USA: 2005.
-
- Nitha B., Meera C.R., Janardhanan K.K. Anti-inflammatory and antitumour activities of cultured mycelium of morel mushroom, Morchella esculenta. Curr. Sci. India. 2007;92:235–239.
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

