High Genomic Diversity and Heterogenous Origins of Pathogenic and Antibiotic-Resistant Escherichia coli in Household Settings Represent a Challenge to Reducing Transmission in Low-Income Settings
- PMID: 31941809
- PMCID: PMC6968650
- DOI: 10.1128/mSphere.00704-19
High Genomic Diversity and Heterogenous Origins of Pathogenic and Antibiotic-Resistant Escherichia coli in Household Settings Represent a Challenge to Reducing Transmission in Low-Income Settings
Abstract
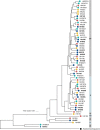
Escherichia coli is present in multiple hosts and environmental compartments as a normal inhabitant, temporary or persistent colonizer, and as a pathogen. Transmission of E. coli between hosts and with the environment is considered to occur more often in areas with poor sanitation. We performed whole-genome comparative analyses on 60 E. coli isolates from soils and fecal sources (cattle, chickens, and humans) in households in rural Bangladesh. Isolates from household soils were in multiple branches of the reconstructed phylogeny, intermixed with isolates from fecal sources. Pairwise differences between all strain pairs were large (minimum, 189 single nucleotide polymorphisms [SNPs]), suggesting high diversity and heterogeneous origins of the isolates. The presence of multiple virulence and antibiotic resistance genes is indicative of the risk that E. coli from soil and feces represent for the transmission of variants that pose potential harm to people. Analysis of the accessory genomes of the Bangladeshi E. coli relative to E. coli genomes available in NCBI identified a common pool of accessory genes shared among E. coli isolates in this geographic area. Together, these findings indicate that in rural Bangladesh, a high level of E. coli in soil is likely driven by contributions from multiple and diverse E. coli sources (human and animal) that share an accessory gene pool relatively unique to previously published E. coli genomes. Thus, interventions to reduce environmental pathogen or antimicrobial resistance transmission should adopt integrated One Health approaches that consider heterogeneous origins and high diversity to improve effectiveness and reduce prevalence and transmission.IMPORTANCEEscherichia coli is reported in high levels in household soil in low-income settings. When E. coli reaches a soil environment, different mechanisms, including survival, clonal expansion, and genetic exchange, have the potential to either maintain or generate E. coli variants with capabilities of causing harm to people. In this study, we used whole-genome sequencing to identify that E. coli isolates collected from rural Bangladeshi household soils, including pathogenic and antibiotic-resistant variants, are diverse and likely originated from multiple diverse sources. In addition, we observed specialization of the accessory genome of this Bangladeshi E. coli compared to E. coli genomes available in current sequence databases. Thus, to address the high level of pathogenic and antibiotic-resistant E. coli transmission in low-income settings, interventions should focus on addressing the heterogeneous origins and high diversity.
Keywords: Escherichia coli; accessory genes; genomic diversity; household settings; soils.
Copyright © 2020 Montealegre et al.
Figures



References
-
- Vila J, Sáez-López E, Johnson JR, Römling U, Dobrindt U, Cantón R, Giske CG, Naas T, Carattoli A, Martínez-Medina M, Bosch J, Retamar P, Rodríguez-Baño J, Baquero F, Soto SM. 2016. Escherichia coli: an old friend with new tidings. FEMS Microbiol Rev 40:437–463. doi: 10.1093/femsre/fuw005. - DOI - PubMed
-
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

