Campylobacter Abundance in Breastfed Infants and Identification of a New Species in the Global Enterics Multicenter Study
- PMID: 31941810
- PMCID: PMC6968651
- DOI: 10.1128/mSphere.00735-19
Campylobacter Abundance in Breastfed Infants and Identification of a New Species in the Global Enterics Multicenter Study
Abstract
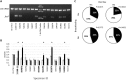
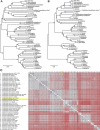
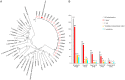
Campylobacter jejuni is a leading cause of bacterial diarrhea worldwide and is associated with high rates of mortality and growth stunting in children inhabiting low- to middle-resource countries. To better understand the impact of breastfeeding on Campylobacter infection in infants in sub-Saharan Africa and South Asia, we examined fecal microbial compositions, bacterial isolates, and their carbohydrate metabolic pathways in Campylobacter-positive infants <1 year of age from the Global Enterics Multicenter Study. Exclusively breastfed infants with diarrhea exhibited high Campylobacter abundances, and this negatively correlated with bacterial carbohydrate metabolism. Although C. jejuni and Campylobacter coli are prevalent among these infants, the second most abundant Campylobacter species was a new species, which we named "Candidatus Campylobacter infans." Asymptomatic Campylobacter carriers also possess significantly different proportions of specific gut microbes compared to diarrheal cases. These findings provide insight into Campylobacter infections in infants in sub-Saharan Africa and South Asia and help inform strategies aimed at eliminating campylobacteriosis in these areas.IMPORTANCECampylobacter is the primary cause of bacterial diarrhea in the United States and can lead to the development of the postinfectious autoimmune neuropathy known as Guillain-Barré syndrome. Also, drug-resistant campylobacters are becoming a serious concern both locally and abroad. In low- and middle-income countries (LMICs), infection with Campylobacter is linked to high rates of morbidity, growth stunting, and mortality in children, and breastfeeding is important for infant nutrition, development, and protection against infectious diseases. In this study, we examined the relationship between breastfeeding and Campylobacter infection and demonstrate the increased selection for C. jejuni and C. coli strains unable to metabolize fucose. We also identify a new Campylobacter species coinfecting these infants with a high prevalence in five of the seven countries in sub-Saharan Africa and South Asia examined. These findings indicate that more detailed studies are needed in LMICs to understand the Campylobacter infection process in order to devise a strategy for eliminating this pathogenic microbe.
Keywords: Campylobacter; GEMS; breastfeeding; l-fucose metabolism; “Candidatus Campylobacter infans,” gut microbiome.
Copyright © 2020 Bian et al.
Figures

References
-
- Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, et al. 2012. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. - DOI - PMC - PubMed
-
- Liu J, Platts-Mills JA, Juma J, Kabir F, Nkeze J, Okoi C, Operario DJ, Uddin J, Ahmed S, Alonso PL, Antonio M, Becker SM, Blackwelder WC, Breiman RF, Faruque ASG, Fields B, Gratz J, Haque R, Hossain A, Hossain MJ, Jarju S, Qamar F, Iqbal NT, Kwambana B, Mandomando I, McMurry TL, Ochieng C, Ochieng JB, Ochieng M, Onyango C, Panchalingam S, Kalam A, Aziz F, Qureshi S, Ramamurthy T, Roberts JH, Saha D, Sow SO, Stroup SE, Sur D, Tamboura B, Taniuchi M, Tennant SM, Toema D, Wu Y, Zaidi A, Nataro JP, Kotloff KL, Levine MM, Houpt ER. 2016. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet 388:1291–1301. doi: 10.1016/S0140-6736(16)31529-X. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
