Intratumoral heterogeneity and clonal evolution in liver cancer
- PMID: 31941899
- PMCID: PMC6962317
- DOI: 10.1038/s41467-019-14050-z
Intratumoral heterogeneity and clonal evolution in liver cancer
Abstract
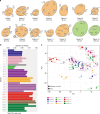
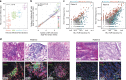
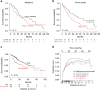
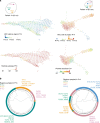
Clonal evolution of a tumor ecosystem depends on different selection pressures that are principally immune and treatment mediated. We integrate RNA-seq, DNA sequencing, TCR-seq and SNP array data across multiple regions of liver cancer specimens to map spatio-temporal interactions between cancer and immune cells. We investigate how these interactions reflect intra-tumor heterogeneity (ITH) by correlating regional neo-epitope and viral antigen burden with the regional adaptive immune response. Regional expression of passenger mutations dominantly recruits adaptive responses as opposed to hepatitis B virus and cancer-testis antigens. We detect different clonal expansion of the adaptive immune system in distant regions of the same tumor. An ITH-based gene signature improves single-biopsy patient survival predictions and an expression survey of 38,553 single cells across 7 regions of 2 patients further reveals heterogeneity in liver cancer. These data quantify transcriptomic ITH and how the different components of the HCC ecosystem interact during cancer evolution.
Conflict of interest statement
A.V. received consulting fees from Guidepoint and Fujifilm; advisory board fees from Exact Sciences, Nucleix and NGM Pharmaceuticals; and lecture fees from Exelixis. J.M.L. is receiving research support from Bayer HealthCare Pharmaceuticals, Eisai Inc, Bristol-Myers Squibb, Boehringer-Ingelheim and Ipsen, and consulting fees from Eli Lilly, Bayer HealthCare Pharmaceuticals, Bristol-Myers Squibb, Eisai Inc, Celsion Corporation, Exelixis, Merck, Ipsen, Roche, Genentech, Glycotest, Navigant, Leerink Swann LLC, Midatech Ltd, Fortress Biotech, Sprink Pharmaceuticals, Nucleix and Can-Fite Biopharma. The remaining authors declare no competing interests.
Figures



References
-
- Amirouchene-Angelozzi Nabil, Swanton Charles, Bardelli Alberto. Tumor Evolution as a Therapeutic Target. Cancer Discovery. 2017;7(8):805–817. doi: 10.1158/2159-8290.CD-17-0343. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

