tRNA 2'-O-methylation by a duo of TRM7/FTSJ1 proteins modulates small RNA silencing in Drosophila
- PMID: 31943105
- PMCID: PMC7038984
- DOI: 10.1093/nar/gkaa002
tRNA 2'-O-methylation by a duo of TRM7/FTSJ1 proteins modulates small RNA silencing in Drosophila
Abstract
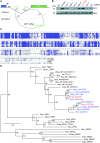
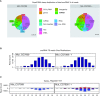
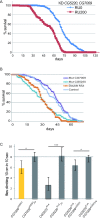
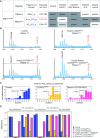
2'-O-Methylation (Nm) represents one of the most common RNA modifications. Nm affects RNA structure and function with crucial roles in various RNA-mediated processes ranging from RNA silencing, translation, self versus non-self recognition to viral defense mechanisms. Here, we identify two Nm methyltransferases (Nm-MTases) in Drosophila melanogaster (CG7009 and CG5220) as functional orthologs of yeast TRM7 and human FTSJ1. Genetic knockout studies together with MALDI-TOF mass spectrometry and RiboMethSeq mapping revealed that CG7009 is responsible for methylating the wobble position in tRNAPhe, tRNATrp and tRNALeu, while CG5220 methylates position C32 in the same tRNAs and also targets additional tRNAs. CG7009 or CG5220 mutant animals were viable and fertile but exhibited various phenotypes such as lifespan reduction, small RNA pathways dysfunction and increased sensitivity to RNA virus infections. Our results provide the first detailed characterization of two TRM7 family members in Drosophila and uncover a molecular link between enzymes catalyzing Nm at specific tRNAs and small RNA-induced gene silencing pathways.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Ayadi L., Galvanin A., Pichot F., Marchand V., Motorin Y.. RNA ribose methylation (2′-O-methylation): occurrence, biosynthesis and biological functions. Biochim. Biophys. Acta Gene Regul. Mech. 2019; 1862:253–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

