Generation of a self-cleaved inducible Cre recombinase for efficient temporal genetic manipulation
- PMID: 31943281
- PMCID: PMC7024834
- DOI: 10.15252/embj.2019102675
Generation of a self-cleaved inducible Cre recombinase for efficient temporal genetic manipulation
Abstract
Site-specific recombinase-mediated genetic technology, such as inducible Cre-loxP recombination (CreER), is widely used for in vivo genetic manipulation with temporal control. The Cre-loxP technology improves our understanding on the in vivo function of specific genes in organ development, tissue regeneration, and disease progression. However, inducible CreER often remains inefficient in gene deletion. In order to improve the efficiency of gene manipulation, we generated a self-cleaved inducible CreER (sCreER) that switches inducible CreER into a constitutively active Cre by itself. We generated endocardial driver Npr3-sCreER and fibroblast driver Col1a2-sCreER, and compared them with conventional Npr3-CreER and Col1a2-CreER, respectively. For easy-to-recombine alleles such as R26-tdTomato, there was no significant difference in recombination efficiency between sCreER and the conventional CreER. However, for alleles that were relatively inert for recombination such as R26-Confetti, R26-LZLT, R26-GFP, or VEGFR2flox/flox alleles, sCreER showed a significantly higher efficiency in recombination compared with conventional CreER in endocardial cells or fibroblasts. Compared with conventional CreER, sCreER significantly enhances the efficiency of recombination to induce gene expression or gene deletion, allowing temporal yet effective in vivo genomic modification for studying gene function in specific cell lineages.
Keywords: Cre-loxP; cell lineages; gene deletion; genetic manipulation.
© 2020 The Authors.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
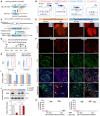
Schematic diagram showing the working principle for sCreER.
Schematic diagram showing steps of switch of sCreER into Cre.
Generation of Npr3‐sCreER or Npr3‐CreER knock‐in alleles by homologous recombination.
Schematic diagram showing the experimental strategy.
Whole‐mount fluorescence view of E14.5‐15.5 hearts from Npr3‐sCreER;R26‐tdTomato and Npr3‐CreER;R26‐tdTomato mice.
Immunostaining for tdTomato, ESR, and CDH5 on heart sections. C.M., compact myocardium; T.M., trabecular myocardium. Arrowheads, tdTomato+ESR− endocardial cells.
Quantification of the percentage of tdTomato+ or ESR+ endothelial cells. Data are mean ± SEM; n = 5.
Isolation of tdTomato− or tdTomato+ cells from E14.5 Npr3‐sCreER;R26‐tdTomato and Npr3‐CreER;R26‐tdTomato mice by FACS.
qRT–PCR of Cre, ESR, Npr3 and Cdh5 from tdTomato+ cells. Data are mean ± SEM; n = 5; *P < 0.05.
Western blotting of ESR and GAPDH in tdTomato− and tdTomato+ cells. Data are mean ± SEM; n = 5; *P < 0.05; n.s., non‐significant.
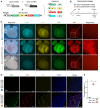
- A
Schematic diagram showing recombination readouts by crossing of Npr3‐CreER or Npr3‐sCreER with R26‐Confetti reporter.
- B
Schematic figure showing experimental strategy.
- C
Whole‐mount fluorescence images of E14.5 Npr3‐CreER;R26‐Confetti and Npr3‐sCreER;R26‐Confetti mouse hearts. No Tam is used as control for leakiness of Npr3‐sCreER.
- D, E
Immunostaining for CDH5 on heart sections shows a significant increase in endothelial cell labeling in Npr3‐sCreER;R26‐Confetti heart compared with Npr3‐CreER;R26‐Confetti heart (Tam). *P < 0.05; data are mean ± SEM; n = 5.
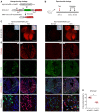
Schematic diagram showing Npr3‐sCreER or Npr3‐CreER mouse crossed with a newly generated Rosa26 reporter R26‐LZLT.
Schematic diagram showing experimental strategy.
Whole‐mount fluorescent images of E14.5 or E15.5 Npr3‐sCreER;R26‐LZLT and Npr3‐CreER;R26‐LZLT mouse hearts. No tam is as control for detecting leakiness of sCreER or CreER.
Representative immunostaining images for tdTomato, ESR, and CDH5 on heart sections.
Quantification of the percentage of endocardial cells expressing tdTomato in Npr3‐sCreER;R26‐LZLT (sCreER) or Npr3‐CreER;R26‐LZLT (CreER) hearts. Data are mean ± SEM; n = 5; P?value was determined using the unpaired Student's t‐test (*P < 0.05).

- A
Schematic diagram showing Kdr gene deletion by sCreER.
- B
Schematic figure showing experimental design.
- C
Immunostaining for VEGFR2 on Npr3‐sCreER;Kdr flox/flox heart section from mice without tamoxifen treatment (No Tam).
- D, E
Immunostaining for VEGFR2 on mouse heart sections after tamoxifen treatment (Tam). A, atrium; V, ventricle; TM, trabecular myocardium.
- F, G
Immunostaining for tdTomato, VEGFR2, and CDH5 on heart sections from mice of four different genotypes. Quantification data showed the percentage of VEGFR2+ endocardial cells (ECs) in tdTomato+ (tdT+) cells in the TM regions. Cartoon images (bottom) indicate tdTomato+ cells (red) and VEGFR2+ cells (green) in the TM or compact myocardium (CM). Data are mean ± SEM; n = 5.
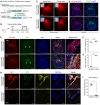
Schematic diagram showing knock‐in of sCreER or CreER into Col1a2 locus by homologous recombination.
Schematic figure showing experimental strategy.
Whole‐mount fluorescence images of Col1a2‐sCreER;R26‐tdTomato or Col1a2‐CreER;R26‐tdTomato hearts. Inserts indicate bright‐field images.
Immunostaining for tdTomato and PDGFRa on heart sections shows similar patterning of tdTomato and PDGFRa signals.
Immunostaining for tdTomato and ESR on heart sections. Arrowheads indicate valve mesenchyme.
Quantification of the percentage of PDGFRa+ cells expressing tdTomato in Col1a2‐sCreER;R26‐tdTomato (sCreER) or Col1a2‐CreER;R26‐tdTomato (CreER) hearts. n.s., non‐significant; data are mean ± SEM; n = 6; n.s., non‐significant.
Quantification of the percentage of tdTomato+ cells expressing ESR. *P < 0.05; data are mean ± SEM; n = 6; *P < 0.05.
Immunostaining for PDGFRa in heart sections shows a significant increase in fibroblast labeling in valves of Col1a2‐sCreER;R26‐Confetti heart compared with Col1a2‐CreER;R26‐Confetti heart (Tam).
Quantification of the percentage of fibroblasts expressing fluorescent reporters in cardiac valves. Data are mean ± SEM; n = 5; *P < 0.05.
References
-
- van Amerongen MJ, Bou‐Gharios G, Popa E, van Ark J, Petersen AH, van Dam GM, van Luyn MJ, Harmsen MC (2008) Bone marrow‐derived myofibroblasts contribute functionally to scar formation after myocardial infarction. J Pathol 214: 377–386 - PubMed
-
- Buckingham ME, Meilhac SM (2011) Tracing cells for tracking cell lineage and clonal behavior. Dev Cell 21: 394–409 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- XDA16010507/Strategic Priority Research Program of the Chinese Academy of Sciences (CAS)/International
- XDB19000000/Strategic Priority Research Program of the Chinese Academy of Sciences (CAS)/International
- 2019YFA0110400/National Key Research & Development Program of China/International
- 2018YFA0107900/National Key Research & Development Program of China/International
- 2018YFA0108100/National Key Research & Development Program of China/International
- 2016YFC1300600/National Key Research & Development Program of China/International
- 2017YFC1001303/National Key Research & Development Program of China/International
- ZJ2018-ZD-004/Shanghai Zhangjiang Stem Cell Project/International
- Youth Innovation Promotion Association of CAS/International
- QYZDB-SSW-SMC003/Key Project of Frontier Sciences of CAS/International
- 19JC1415700/Shanghai Science and Technology Commission/International
- 17ZR1449600/Shanghai Science and Technology Commission/International
- 17ZR1449800/Shanghai Science and Technology Commission/International
- 2017ZT07S347/Program for Guangdong Introduction Innovative and Entrepreneurial Teams/International
- ZJ2018-ZD-004/Major Program of Development Fund for Shanghai Zhangjiang National Innovation Demonstration Zone (Stem Cell Strategic Biobank and Stem Cell Clinical Technology Transformation Platform)/International
- Shanghai Yangfan Project/International
- China Postdoctoral Innovative Talent Support Program/International
- 2018QNRC001/CAST/International
- 2017QNRC001/CAST/International
- Boehringer Ingelheim/International
- Sanofi-SIBS Fellowship/International
- Royal Society-Newton Advanced Fellowship/International
- 31730112/National Science Foundation of China/International
- 91639302/National Science Foundation of China/International
- 31625019/National Science Foundation of China/International
- 91849202/National Science Foundation of China/International
- 81761138040/National Science Foundation of China/International
- 81872241/National Science Foundation of China/International
- 31701292/National Science Foundation of China/International
- 31801215/National Science Foundation of China/International
- 31922032/National Science Foundation of China/International
- China Postdoctoral Science Foundation/International
- AstraZeneca/International
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

