Explainable Anatomical Shape Analysis Through Deep Hierarchical Generative Models
- PMID: 31944949
- PMCID: PMC7269693
- DOI: 10.1109/TMI.2020.2964499
Explainable Anatomical Shape Analysis Through Deep Hierarchical Generative Models
Abstract
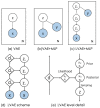
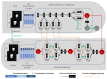
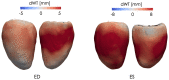
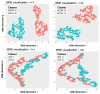
Quantification of anatomical shape changes currently relies on scalar global indexes which are largely insensitive to regional or asymmetric modifications. Accurate assessment of pathology-driven anatomical remodeling is a crucial step for the diagnosis and treatment of many conditions. Deep learning approaches have recently achieved wide success in the analysis of medical images, but they lack interpretability in the feature extraction and decision processes. In this work, we propose a new interpretable deep learning model for shape analysis. In particular, we exploit deep generative networks to model a population of anatomical segmentations through a hierarchy of conditional latent variables. At the highest level of this hierarchy, a two-dimensional latent space is simultaneously optimised to discriminate distinct clinical conditions, enabling the direct visualisation of the classification space. Moreover, the anatomical variability encoded by this discriminative latent space can be visualised in the segmentation space thanks to the generative properties of the model, making the classification task transparent. This approach yielded high accuracy in the categorisation of healthy and remodelled left ventricles when tested on unseen segmentations from our own multi-centre dataset as well as in an external validation set, and on hippocampi from healthy controls and patients with Alzheimer's disease when tested on ADNI data. More importantly, it enabled the visualisation in three-dimensions of both global and regional anatomical features which better discriminate between the conditions under exam. The proposed approach scales effectively to large populations, facilitating high-throughput analysis of normal anatomy and pathology in large-scale studies of volumetric imaging.
Figures




References
-
- Attar R, et al. Quantitative CMR Population imaging on 20,000 subjects of the UK Biobank imaging study: LV/RV quantification pipeline and its evaluation. Medical Image Analysis. 2019 - PubMed
-
- Nogovitsyn N, et al. Testing a deep convolutional neural network for automated hippocampus segmentation in a longitudinal sample of healthy participants. NeuroImage. 2019;197:589–597. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

