Comparing bioinformatic pipelines for microbial 16S rRNA amplicon sequencing
- PMID: 31945086
- PMCID: PMC6964864
- DOI: 10.1371/journal.pone.0227434
Comparing bioinformatic pipelines for microbial 16S rRNA amplicon sequencing
Abstract
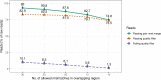
Microbial amplicon sequencing studies are an important tool in biological and biomedical research. Widespread 16S rRNA gene microbial surveys have shed light on the structure of many ecosystems inhabited by bacteria, including the human body. However, specialized software and algorithms are needed to convert raw sequencing data into biologically meaningful information (i.e. tables of bacterial counts). While different bioinformatic pipelines are available in a rapidly changing and improving field, users are often unaware of limitations and biases associated with individual pipelines and there is a lack of agreement regarding best practices. Here, we compared six bioinformatic pipelines for the analysis of amplicon sequence data: three OTU-level flows (QIIME-uclust, MOTHUR, and USEARCH-UPARSE) and three ASV-level (DADA2, Qiime2-Deblur, and USEARCH-UNOISE3). We tested workflows with different quality control options, clustering algorithms, and cutoff parameters on a mock community as well as on a large (N = 2170) recently published fecal sample dataset from the multi-ethnic HELIUS study. We assessed the sensitivity, specificity, and degree of consensus of the different outputs. DADA2 offered the best sensitivity, at the expense of decreased specificity compared to USEARCH-UNOISE3 and Qiime2-Deblur. USEARCH-UNOISE3 showed the best balance between resolution and specificity. OTU-level USEARCH-UPARSE and MOTHUR performed well, but with lower specificity than ASV-level pipelines. QIIME-uclust produced large number of spurious OTUs as well as inflated alpha-diversity measures and should be avoided in future studies. This study provides guidance for researchers using amplicon sequencing to gain biological insights.
Conflict of interest statement
E.L is employed by Horaizon BV. Horaizon BV did not play any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript and only provided financial support in the form of authors' salaries.This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

