Pan-cancer analysis of somatic mutations and epigenetic alterations in insulated neighbourhood boundaries
- PMID: 31945090
- PMCID: PMC6964824
- DOI: 10.1371/journal.pone.0227180
Pan-cancer analysis of somatic mutations and epigenetic alterations in insulated neighbourhood boundaries
Abstract
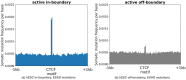
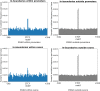
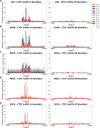
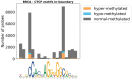
Recent evidence shows that the disruption of constitutive insulated neighbourhoods might lead to oncogene dysregulation. We present here a systematic pan-cancer characterisation of the associations between constitutive boundaries and genome alterations in cancer. Specifically, we investigate the enrichment of somatic mutation, abnormal methylation, and copy number alteration events in the proximity of CTCF bindings overlapping with topological boundaries (junctions) in 26 cancer types. Focusing on CTCF motifs that are both in-boundary (overlapping with junctions) and active (overlapping with peaks of CTCF expression), we find a significant enrichment of somatic mutations in several cancer types. Furthermore, mutated junctions are significantly conserved across cancer types, and we also observe a positive selection of transversions rather than transitions in many cancer types. We also analyzed the mutational signature found on the different classes of CTCF motifs, finding some signatures (such as SBS26) to have a higher weight within in-boundary than off-bounday motifs. Regarding methylation, we find a significant number of over-methylated active in-boundary CTCF motifs in several cancer types; similarly to somatic-mutated junctions, they also have a significant conservation across cancer types. Finally, in several cancer types we observe that copy number alterations tend to overlap with active junctions more often than in matched normal samples. While several articles have recently reported a mutational enrichment at CTCF binding sites for specific cancer types, our analysis is pan-cancer and investigates abnormal methylation and copy number alterations in addition to somatic mutations. Our method is fully replicable and suggests several follow-up tumour-specific analyses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

