Regional adaptations and parallel mutations in Feline panleukopenia virus strains from China revealed by nearly-full length genome analysis
- PMID: 31945103
- PMCID: PMC6964837
- DOI: 10.1371/journal.pone.0227705
Regional adaptations and parallel mutations in Feline panleukopenia virus strains from China revealed by nearly-full length genome analysis
Abstract
Protoparvoviruses, widespread among cats and wild animals, are responsible for leukopenia. Feline panleukopenia virus (FPLV) in domestic cats is genetically diverse and some strains may differ from those used for vaccination. The presence of FPLV in two domestic cats from Hebei Province in China was identified by polymerase chain reaction. Samples from these animals were used to isolate FPLV strains in CRFK cells for genome sequencing. Phylogenetic analysis was performed to compare our isolates with available sequences of FPLV, mink parvovirus (MEV) and canine parvovirus (CPV). The isolated strains were closely related to strains of FPLV/MEV isolated in the 1960s. Our analysis also revealed that the evolutionary history of FPLV and MEV is characterized by local adaptations in the Vp2 gene. Thus, it is likely that new FPLV strains are emerging to evade the anti-FPLV immune response.
Conflict of interest statement
All the authors have declared that no competing interests exist.
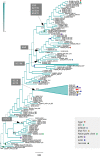
Figures
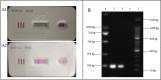


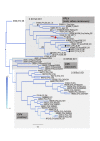

Similar articles
-
Differences in the evolutionary pattern of feline panleukopenia virus and canine parvovirus.Virology. 1998 Sep 30;249(2):440-52. doi: 10.1006/viro.1998.9335. Virology. 1998. PMID: 9791034
-
Genetic analysis of feline panleukopenia viruses from cats with gastroenteritis.J Gen Virol. 2008 Sep;89(Pt 9):2290-2298. doi: 10.1099/vir.0.2008/001503-0. J Gen Virol. 2008. PMID: 18753239
-
Molecular analysis of partial VP-2 gene amplified from rectal swab samples of diarrheic dogs in Pakistan confirms the circulation of canine parvovirus genetic variant CPV-2a and detects sequences of feline panleukopenia virus (FPV).Virol J. 2018 Mar 15;15(1):45. doi: 10.1186/s12985-018-0958-y. Virol J. 2018. PMID: 29544546 Free PMC article.
-
Feline panleukopenia virus: its interesting evolution and current problems in immunoprophylaxis against a serious pathogen.Vet Microbiol. 2013 Jul 26;165(1-2):29-32. doi: 10.1016/j.vetmic.2013.02.005. Epub 2013 Feb 18. Vet Microbiol. 2013. PMID: 23561891 Review.
-
Parvovirus infection in domestic companion animals.Vet Clin North Am Small Anim Pract. 2008 Jul;38(4):837-50, viii-ix. doi: 10.1016/j.cvsm.2008.03.008. Vet Clin North Am Small Anim Pract. 2008. PMID: 18501282 Review.
Cited by
-
Small but mighty: old and new parvoviruses of veterinary significance.Virol J. 2021 Oct 24;18(1):210. doi: 10.1186/s12985-021-01677-y. Virol J. 2021. PMID: 34689822 Free PMC article. Review.
-
Novel Chaphamaparvovirus in Insectivorous Molossus molossus Bats, from the Brazilian Amazon Region.Viruses. 2023 Feb 22;15(3):606. doi: 10.3390/v15030606. Viruses. 2023. PMID: 36992315 Free PMC article.
-
Natural infection of parvovirus in wild fishing cats (Prionailurus viverrinus) reveals extant viral localization in kidneys.PLoS One. 2021 Mar 2;16(3):e0247266. doi: 10.1371/journal.pone.0247266. eCollection 2021. PLoS One. 2021. PMID: 33651823 Free PMC article.
-
First detection and complete genome analysis of the Lyon IARC polyomavirus in China from samples of diarrheic cats.Virus Genes. 2021 Jun;57(3):284-288. doi: 10.1007/s11262-021-01840-1. Epub 2021 May 10. Virus Genes. 2021. PMID: 33970402 Free PMC article.
-
Identification of a Feline Panleukopenia Virus from Captive Giant Pandas (Ailuropoda melanoleuca) and Its Phylogenetic Analysis.Transbound Emerg Dis. 2023 May 19;2023:7721487. doi: 10.1155/2023/7721487. eCollection 2023. Transbound Emerg Dis. 2023. PMID: 40303714 Free PMC article.
References
-
- Decaro N.; Camero M.; Greco G.; Zizzo N.; Tinelli A.; Campolo M.; et al. Canine distemper and related diseases: Report of a severe outbreak in a kennel. New Microbiol 2004, 27, 177–181. - PubMed
-
- Kariatsumari T.; Horiuchi M.; Hama E.; Yaguchi K.; Ishigurio N.; Goto H.; et al. Construction and nucleotide sequence analysis of an infectious DNA clone of the autonomous parvovirus, mink enteritis virus. J Gen Virol 1991, 72 (Pt 4), 867–875. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

