Cytoplasmic factories, virus assembly, and DNA replication kinetics collectively constrain the formation of poxvirus recombinants
- PMID: 31945138
- PMCID: PMC6964908
- DOI: 10.1371/journal.pone.0228028
Cytoplasmic factories, virus assembly, and DNA replication kinetics collectively constrain the formation of poxvirus recombinants
Abstract
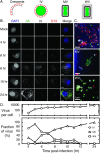
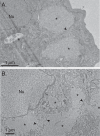
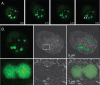
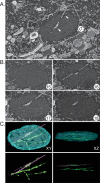
Poxviruses replicate in cytoplasmic structures called factories and each factory begins as a single infecting particle. Sixty-years ago Cairns predicted that this might have effects on vaccinia virus (VACV) recombination because the factories would have to collide and mix their contents to permit recombination. We've since shown that factories collide irregularly and that even then the viroplasm mixes poorly. We've also observed that while intragenic recombination occurs frequently early in infection, intergenic recombination is less efficient and happens late in infection. Something inhibits factory fusion and viroplasm mixing but what is unclear. To study this, we've used optical and electron microscopy to track factory movement in co-infected cells and correlate these observations with virus development and recombinant formation. While the technical complexity of the experiments limited the number of cells that are amenable to extensive statistical analysis, these studies do show that intergenic recombination coincides with virion assembly and when VACV replication has declined to ≤10% of earlier levels. Along the boundaries between colliding factories, one sees ER membrane remnants and other cell constituents like mitochondria. These collisions don't always cause factory fusion, but when factories do fuse, they still entrain cell constituents like mitochondria and ER-wrapped microtubules. However, these materials wouldn't seem to pose much of a further barrier to DNA mixing and so it's likely that the viroplasm also presents an omnipresent impediment to DNA mixing. Late packaging reactions might help to disrupt the viroplasm, but packaging would sequester the DNA just as the replication and recombination machinery goes into decline and further reduce recombinant yields. Many factors thus appear to conspire to limit recombination between co-infecting poxviruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

