Codon Directional Asymmetry Suggests Swapped Prebiotic 1st and 2nd Codon Positions
- PMID: 31948054
- PMCID: PMC6981979
- DOI: 10.3390/ijms21010347
Codon Directional Asymmetry Suggests Swapped Prebiotic 1st and 2nd Codon Positions
Abstract
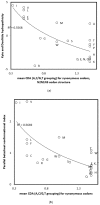
Background: Codon directional asymmetry (CDA) classifies the 64 codons into palindromes (XYX, CDA = 0), and 5'- and 3'-dominant (YXX and XXY, CDA < 0 and CDA > 0, respectively). Previously, CDA was defined by the purine/pyrimidine divide (A,G/C,T), where X is either a purine or a pyrimidine. For the remaining codons with undefined CDA, CDA was defined by the 5' or 3' nucleotide complementary to Y. This CDA correlates with cognate amino acid tRNA synthetase classes, antiparallel beta sheet conformation index and the evolutionary order defined by the self-referential genetic code evolution model (CDA < 0: class I, high beta sheet index, late genetic code inclusion). Methods: We explore associations of CDAs defined by nucleotide classifications according to complementarity strengths (A:T, weak; C:G, strong) and keto-enol/amino-imino groupings (G,T/A,C), also after swapping 1st and 2nd codon positions with amino acid physicochemical and structural properties. Results: Here, analyses show that for the eight codons whose purine/pyrimidine-based CDA requires using the rule of complementarity with the midposition, using weak interactions to define CDA instead of complementarity increases associations with tRNA synthetase classes, antiparallel beta sheet index and genetic code evolutionary order. CDA defined by keto-enol/amino-imino groups, 1st and 2nd codon positions swapped, correlates with amino acid parallel beta sheet formation indices and Doolittle's hydropathicities. Conclusions: Results suggest (a) prebiotic swaps from N2N1N3 to N1N2N3 codon structures, (b) that tRNA-mediated translation replaced direct codon-amino acid interactions, and (c) links between codon structures and cognate amino acid properties.
Keywords: amino-imino tautomer; anticodon; bijective transformation; keto-enol tautomer; swinger RNA; systematic nucleotide deletion; systematic nucleotide exchange.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Bias for 3'-Dominant Codon Directional Asymmetry in Theoretical Minimal RNA Rings.J Comput Biol. 2019 Sep;26(9):1003-1012. doi: 10.1089/cmb.2018.0256. Epub 2019 May 22. J Comput Biol. 2019. PMID: 31120344
-
Deamination gradients within codons after 1<->2 position swap predict amino acid hydrophobicity and parallel β-sheet conformational preference.Biosystems. 2020 May;191-192:104116. doi: 10.1016/j.biosystems.2020.104116. Epub 2020 Feb 18. Biosystems. 2020. PMID: 32081715
-
Genetic Code Optimization for Cotranslational Protein Folding: Codon Directional Asymmetry Correlates with Antiparallel Betasheets, tRNA Synthetase Classes.Comput Struct Biotechnol J. 2017 Aug 12;15:412-424. doi: 10.1016/j.csbj.2017.08.001. eCollection 2017. Comput Struct Biotechnol J. 2017. PMID: 28924459 Free PMC article.
-
Maximal Genetic Code Symmetry Is a Physicochemical Purine-Pyrimidine Symmetry Language for Transcription and Translation in the Flow of Genetic Information from DNA to Proteins.Int J Mol Sci. 2024 Sep 2;25(17):9543. doi: 10.3390/ijms25179543. Int J Mol Sci. 2024. PMID: 39273490 Free PMC article. Review.
-
Evolution of the aminoacyl-tRNA synthetases and the origin of the genetic code.J Mol Evol. 1995 May;40(5):545-50. doi: 10.1007/BF00166624. J Mol Evol. 1995. PMID: 7783229 Review.
Cited by
-
The primordial tRNA acceptor stem code from theoretical minimal RNA ring clusters.BMC Genet. 2020 Jan 23;21(1):7. doi: 10.1186/s12863-020-0812-2. BMC Genet. 2020. PMID: 31973715 Free PMC article.
References
-
- Van Valen L. A study of fluctuating asymmetry. Evolution. 1962;16:125–142. doi: 10.1111/j.1558-5646.1962.tb03206.x. - DOI
-
- Palmer A.R. Fluctuating asymmetry analyses: A primer. In: Markow T.A., editor. Developmental Instability: Its Origins and Evolutionary Implications. Kluwer Academic Publishers; Dordrecht, the Netherlands: 1994. pp. 335–364.
-
- Werner Y.L., Rothenstein D., Sivan N. Directional asymmetry in reptiles (Sauria, Gekkonidae—Ptyodactylus) and its possible evolutionarty role, with implications for biometrical methods. J. Zool. 1991;225:647–658. doi: 10.1111/j.1469-7998.1991.tb04331.x. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous