Systematic Analysis of the Maize OSCA Genes Revealing ZmOSCA Family Members Involved in Osmotic Stress and ZmOSCA2.4 Confers Enhanced Drought Tolerance in Transgenic Arabidopsis
- PMID: 31948071
- PMCID: PMC6982122
- DOI: 10.3390/ijms21010351
Systematic Analysis of the Maize OSCA Genes Revealing ZmOSCA Family Members Involved in Osmotic Stress and ZmOSCA2.4 Confers Enhanced Drought Tolerance in Transgenic Arabidopsis
Abstract
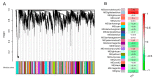
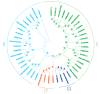
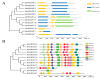
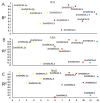
OSCAs are hyperosmolality-gated calcium-permeable channel proteins. In this study, two co-expression modules, which are strongly associated with maize proline content, were screened by weighted correlation network analysis, including three ZmOSCA family members. Phylogenetic and protein domain analyses revealed that 12 ZmOSCA members were classified into four classes, which all contained DUF221 domain. The promoter region contained multiple core elements responsive to abiotic stresses and hormones. Colinear analysis revealed that ZmOSCAs had diversified prior to maize divergence. Most ZmOSCAs responded positively to ABA, PEG, and NaCl treatments. ZmOSCA2.3 and ZmOSCA2.4 were up-regulated by more than 200-fold under the three stresses, and showed significant positive correlations with proline content. Yeast two-hybrid and bimolecular fluorescence complementation indicated that ZmOSCA2.3 and ZmOSCA2.4 proteins interacted with ZmEREB198. Over-expression of ZmOSCA2.4 in Arabidopsis remarkably improved drought resistance. Moreover, over-expression of ZmOSCA2.4 enhanced the expression of drought tolerance-associated genes and reduced the expression of senescence-associated genes. We also found that perhaps ZmOSCA2.4 was regulated by miR5054.The results provide a high-quality molecular resource for selecting resistant breeding, and lay a foundation for elucidating regulatory mechanism of ZmOSCA under abiotic stresses.
Keywords: DUF221 domain; OSCAs; abiotic stresses; co-expression modules; drought resistance; interaction; proline content.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Dornhorst A.C. Review of Medical Physiology. Anesthesiology. 2001;52:959–960.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

