LncRNA NEAT1 affects inflammatory response by targeting miR-129-5p and regulating Notch signaling pathway in epilepsy
- PMID: 31948324
- PMCID: PMC7100884
- DOI: 10.1080/15384101.2020.1711578
LncRNA NEAT1 affects inflammatory response by targeting miR-129-5p and regulating Notch signaling pathway in epilepsy
Abstract
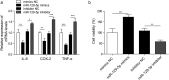
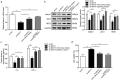
It is crucial to understand the molecular mechanisms involved in epileptogenesis. This study aims to investigate the role of lncRNA NEAT1, miR-129-5p and Notch signaling pathway in epilepsy. In this research, temporal lobe tissues were collected from patients with epilepsy and healthy controls. The CTX-TNA cells were treated with IL-1β to establis
Keywords: Epilepsy; LncRNA NEAT1; MiR-129-5p; Notch signaling pathway.
Figures





Similar articles
-
Deficiency of NEAT1 prevented MPP+-induced inflammatory response, oxidative stress and apoptosis in dopaminergic SK-N-SH neuroblastoma cells via miR-1277-5p/ARHGAP26 axis.Brain Res. 2021 Jan 1;1750:147156. doi: 10.1016/j.brainres.2020.147156. Epub 2020 Oct 16. Brain Res. 2021. PMID: 33069733
-
Long Non-Coding RNA Nuclear Paraspeckle Assembly Transcript 1 (NEAT1)Relieves Sepsis-Induced Kidney Injury and Lipopolysaccharide (LPS)-Induced Inflammation in HK-2 Cells.Med Sci Monit. 2020 Jul 29;26:e921906. doi: 10.12659/MSM.921906. Med Sci Monit. 2020. PMID: 32724027 Free PMC article.
-
Long non-coding RNA NEAT1 promotes lipopolysaccharide-induced injury in human tubule epithelial cells by regulating miR-93-5p/TXNIP axis.Med Microbiol Immunol. 2021 Jun;210(2-3):121-132. doi: 10.1007/s00430-021-00705-6. Epub 2021 Apr 22. Med Microbiol Immunol. 2021. PMID: 33885954
-
Long noncoding RNA SNHG16 targets miR-146a-5p/CCL5 to regulate LPS-induced WI-38 cell apoptosis and inflammation in acute pneumonia.Life Sci. 2019 Jul 1;228:189-197. doi: 10.1016/j.lfs.2019.05.008. Epub 2019 May 7. Life Sci. 2019. PMID: 31071307 Review.
-
The interaction between miRNAs/lncRNAs and Notch pathway in human disorders.Biomed Pharmacother. 2021 Jun;138:111496. doi: 10.1016/j.biopha.2021.111496. Epub 2021 Mar 17. Biomed Pharmacother. 2021. PMID: 33743335 Review.
Cited by
-
LINC01614 Promotes Colorectal Cancer Cell Growth and Migration by Regulating miR-217-5p/HMGA1 Axis.Anal Cell Pathol (Amst). 2023 May 31;2023:6833987. doi: 10.1155/2023/6833987. eCollection 2023. Anal Cell Pathol (Amst). 2023. PMID: 39282156 Free PMC article.
-
LncRNA ZNF883-Mediated NLRP3 Inflammasome Activation and Epilepsy Development Involve USP47 Upregulation.Mol Neurobiol. 2022 Aug;59(8):5207-5221. doi: 10.1007/s12035-022-02902-7. Epub 2022 Jun 9. Mol Neurobiol. 2022. PMID: 35678979
-
Dexamethasone Alleviates Myocardial Injury in a Rat Model of Acute Myocardial Infarction Supported by Venoarterial Extracorporeal Membrane Oxygenation.Front Public Health. 2022 Jul 19;10:900751. doi: 10.3389/fpubh.2022.900751. eCollection 2022. Front Public Health. 2022. PMID: 35928492 Free PMC article.
-
Long non-coding RNAs: Potential therapeutic targets for epilepsy.Front Neurosci. 2022 Oct 6;16:986874. doi: 10.3389/fnins.2022.986874. eCollection 2022. Front Neurosci. 2022. PMID: 36278003 Free PMC article. Review.
-
CircINTS4 Facilitates Chemoresistance of TNBC by Competitively Binding miR-129-5p/POM121 Axis.J Oncol. 2022 Apr 4;2022:2630864. doi: 10.1155/2022/2630864. eCollection 2022. J Oncol. 2022. PMID: 35419056 Free PMC article.
References
-
- Camfield P, Camfield C.. Regression in children with epilepsy. Neurosci Biobehav Rev. 2019;96:210–218. - PubMed
-
- Huang WS, Zhu L.. MiR-134 expression and changes in inflammatory cytokines of rats with epileptic seizures. Eur Rev Med Pharmacol Sci. 2018;22(11):3479–3484. - PubMed
-
- Wang HK, Yan H, Wang K, et al. Dynamic regulation effect of long non-coding RNA-UCA1 on NF-kB in hippocampus of epilepsy rats. Eur Rev Med Pharmacol Sci. 2017;21(13):3113–3119. - PubMed
-
- Aronica E, Ravizza T, Zurolo E, et al. Astrocyte immune responses in epilepsy. Glia. 2012;60(8):1258–1268. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
