Lag penalized weighted correlation for time series clustering
- PMID: 31948388
- PMCID: PMC6966853
- DOI: 10.1186/s12859-019-3324-1
Lag penalized weighted correlation for time series clustering
Abstract
Background: The similarity or distance measure used for clustering can generate intuitive and interpretable clusters when it is tailored to the unique characteristics of the data. In time series datasets generated with high-throughput biological assays, measurements such as gene expression levels or protein phosphorylation intensities are collected sequentially over time, and the similarity score should capture this special temporal structure.
Results: We propose a clustering similarity measure called Lag Penalized Weighted Correlation (LPWC) to group pairs of time series that exhibit closely-related behaviors over time, even if the timing is not perfectly synchronized. LPWC aligns time series profiles to identify common temporal patterns. It down-weights aligned profiles based on the length of the temporal lags that are introduced. We demonstrate the advantages of LPWC versus existing time series and general clustering algorithms. In a simulated dataset based on the biologically-motivated impulse model, LPWC is the only method to recover the true clusters for almost all simulated genes. LPWC also identifies clusters with distinct temporal patterns in our yeast osmotic stress response and axolotl limb regeneration case studies.
Conclusions: LPWC achieves both of its time series clustering goals. It groups time series with correlated changes over time, even if those patterns occur earlier or later in some of the time series. In addition, it refrains from introducing large shifts in time when searching for temporal patterns by applying a lag penalty. The LPWC R package is available at https://github.com/gitter-lab/LPWC and CRAN under a MIT license.
Keywords: Hierarchical clustering; Temporal alignment; Unsupervised learning.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




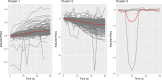

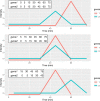
References
-
- Bar-Joseph Z, Gitter A, Simon I. Studying and modelling dynamic biological processes using time-series gene expression data. Nat Rev Genet. 2012;13(8):552–64. - PubMed
-
- Liang Y, Kelemen A. Dynamic modeling and network approaches for omics time course data: overview of computational approaches and applications. Brief Bioinform. 2017. 10.1093/bib/bbx036. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

