The Many Models of Strigolactone Signaling
- PMID: 31948791
- PMCID: PMC7184880
- DOI: 10.1016/j.tplants.2019.12.009
The Many Models of Strigolactone Signaling
Abstract
Strigolactones (SLs) are a class of plant hormones involved in several biological processes that are of great agricultural concern. While initiating plant-fungal symbiosis, SLs also trigger germination of parasitic plants that pose a major threat to farming. In vascular plants, SLs control shoot branching, which is linked to crop yield. SL research has been a fascinating field that has produced a variety of different signaling models, reflecting a complex picture of hormone perception. Here, we review recent developments in the SL field and the crystal structures that gave rise to various models of receptor activation. We also highlight the increasing number of discovered SL molecules, reflecting the existence of cross-kingdom SL communication.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Figures
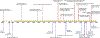


References
-
- Cook CE. et al. (1966) Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154, 1189–1190 - PubMed
-
- Akiyama K. et al. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827 - PubMed
-
- Proust H. et al. (2011) Strigolactones regulate protonema branching and act as a quorum sensing-like signal in the moss Physcomitrella patens. Development 138, 1531–1539 - PubMed
-
- Gomez-Roldan V. et al. (2008) Strigolactone inhibition of shoot branching. Nature 455, 189–194 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

