Receptor-mediated cell entry of paramyxoviruses: Mechanisms, and consequences for tropism and pathogenesis
- PMID: 31949044
- PMCID: PMC7049954
- DOI: 10.1074/jbc.REV119.009961
Receptor-mediated cell entry of paramyxoviruses: Mechanisms, and consequences for tropism and pathogenesis
Abstract
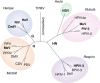
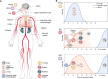
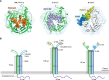
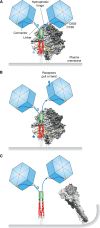
Research in the last decade has uncovered many new paramyxoviruses, airborne agents that cause epidemic diseases in animals including humans. Most paramyxoviruses enter epithelial cells of the airway using sialic acid as a receptor and cause only mild disease. However, others cross the epithelial barrier and cause more severe disease. For some of these viruses, the host receptors have been identified, and the mechanisms of cell entry have been elucidated. The tetrameric attachment proteins of paramyxoviruses have vastly different binding affinities for their cognate receptors, which they contact through different binding surfaces. Nevertheless, all input signals are converted to the same output: conformational changes that trigger refolding of trimeric fusion proteins and membrane fusion. Experiments with selectively receptor-blinded viruses inoculated into their natural hosts have provided insights into tropism, identifying the cells and tissues that support growth and revealing the mechanisms of pathogenesis. These analyses also shed light on diabolically elegant mechanisms used by morbilliviruses, including the measles virus, to promote massive amplification within the host, followed by efficient aerosolization and rapid spread through host populations. In another paradigm of receptor-facilitated severe disease, henipaviruses, including Nipah and Hendra viruses, use different members of one protein family to cause zoonoses. Specific properties of different paramyxoviruses, like neurotoxicity and immunosuppression, are now understood in the light of receptor specificity. We propose that research on the specific receptors for several newly identified members of the Paramyxoviridae family that may not bind sialic acid is needed to anticipate their zoonotic potential and to generate effective vaccines and antiviral compounds.
Keywords: Nipah virus; animal virus; cell invasion; host-pathogen interaction; infection; measles; microbiology; morbillivirus; negative-strand RNA virus; paramyxovirus; pathogenesis; receptor; sialic acid; virology; virus entry.
© 2020 Navaratnarajah et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
-
- Lamb R. A., and Parks G. (eds) (2013) Paramyxoviridae, Vol. 1, pp. 957–995, Lippincott Williams & Wilkins, Philadelphia
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

