Landscape of transcriptome variations uncovering known and novel driver events in colorectal carcinoma
- PMID: 31949199
- PMCID: PMC6965099
- DOI: 10.1038/s41598-019-57311-z
Landscape of transcriptome variations uncovering known and novel driver events in colorectal carcinoma
Abstract
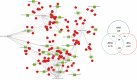
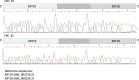
We focused on an integrated view of genomic changes in Colorectal cancer (CRC) and distant normal colon tissue (NTC) to test the effectiveness of expression profiling on identification of molecular targets. We performed transcriptome on 16 primary coupled CRC and NTC tissues. We identified pathways and networks related to pathophysiology of CRC and selected potential therapeutic targets. CRC cells have multiple ways to reprogram its transcriptome: a functional enrichment analysis in 285 genes, 25% mutated, showed that they control the major cellular processes known to promote tumorigenesis. Among the genes showing alternative splicing, cell cycle related genes were upregulated (CCND1, CDC25B, MCM2, MCM3), while genes involved in fatty acid metabolism (ACAAA2, ACADS, ACAT1, ACOX, CPT1A, HMGCS2) were downregulated. Overall 148 genes showed differential splicing identifying 17 new isoforms. Most of them are involved in the pathogenesis of CRC, although the functions of these variants remain unknown. We identified 2 in-frame fusion events, KRT19-KRT18 and EEF1A1-HSP90AB1, encoding for chemical proteins in two CRC patients. We draw a functional interactome map involving integrated multiple genomic features in CRC. Finally, we underline that two functional cell programs are prevalently deregulated and absolutely crucial to determinate and sustain CRC phenotype.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

