Fam151b, the mouse homologue of C.elegans menorin gene, is essential for retinal function
- PMID: 31949211
- PMCID: PMC6965129
- DOI: 10.1038/s41598-019-57398-4
Fam151b, the mouse homologue of C.elegans menorin gene, is essential for retinal function
Abstract
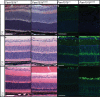
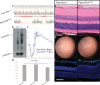
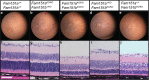
Fam151b is a mammalian homologue of the C. elegans menorin gene, which is involved in neuronal branching. The International Mouse Phenotyping Consortium (IMPC) aims to knock out every gene in the mouse and comprehensively phenotype the mutant animals. This project identified Fam151b homozygous knock-out mice as having retinal degeneration. We show they have no photoreceptor function from eye opening, as demonstrated by a lack of electroretinograph (ERG) response. Histological analysis shows that during development of the eye the correct number of cells are produced and that the layers of the retina differentiate normally. However, after eye opening at P14, Fam151b mutant eyes exhibit signs of retinal stress and rapidly lose photoreceptor cells. We have mutated the second mammalian menorin homologue, Fam151a, and homozygous mutant mice have no discernible phenotype. Sequence analysis indicates that the FAM151 proteins are members of the PLC-like phosphodiesterase superfamily. However, the substrates and function of the proteins remains unknown.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Phelan JK, Bok D. A brief review of retinitis pigmentosa and the identified retinitis pigmentosa genes. Mol Vis. 2000;6:24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

