DNA sensors, crucial receptors to resist pathogens, are deregulated in colorectal cancer and associated with initiation and progression of the disease
- PMID: 31949493
- PMCID: PMC6959017
- DOI: 10.7150/jca.34188
DNA sensors, crucial receptors to resist pathogens, are deregulated in colorectal cancer and associated with initiation and progression of the disease
Abstract
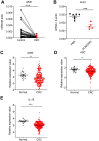
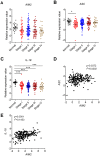
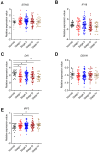
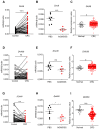
Background: DNA sensors are innate immune receptors that detect intracellular endogenous or exogenous DNA. They are critical to trigger immune response against DNA viral and intracellular bacterial infection, and are involved in inflammatory diseases and tumorigenesis. Recent accumulating evidences indicated that DNA sensors are also crucial for controlling the development of colorectal cancer (CRC). However, a systematic study on the expression profile of DNA sensors in CRC and their clinical significance are still lacking. Methods: We investigated the expression profile of DNA sensors in CRC and their clinical significance by taking advantage of clinical CRC samples, mouse AOM/DSS treatment model, and Oncomine ® bioinformatics platform. Results: Our study identified that the expression of DNA sensors, including AIM2, DAI, as well as inflammasome molecules ASC/IL-18, TLR9 and adaptor MyD88, and DDX60 decreased in human CRC, whereas the expression of DHX9, DHX36, and DDX41 significantly increased. Among them, the expression of AIM2/ASC/IL-18, MyD88, DAI, DHX36, and DDX60 were associated with cancer stages. In addition, we also performed correlation analysis between DNA sensors and their main signaling molecules to explore the possible mechanisms. The results showed that there were positive correlations between AIM2 and ASC/IL-18, DHX9 and MAVS, and TLR9 and MyD88 expression. In addition, the gene expression patterns of some DNA sensors were confirmed by Western-blot analysis. Conclusions: Our study revealed that the expression of multiple DNA sensors was deregulated in CRC and might be involved in tumor development. More importantly, the study identified that, among all these DNA sensors, AIM2, DAI, and DDX60 could be potentially critical for diagnosis, prognosis, and therapy of CRC and deserve further investigation.
Keywords: DNA sensors; cancer; cancer stages; expression profile.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–91. - PubMed
-
- Jess T, Rungoe C, Peyrin-Biroulet L. Risk of colorectal cancer in patients with ulcerative colitis: a meta-analysis of population-based cohort studies. Clin Gastroenterol Hepatol. 2012;10:639–45. - PubMed
-
- Wang K, Karin M. Tumor-Elicited Inflammation and Colorectal Cancer. Adv Cancer Res. 2015;128:173–96. - PubMed
-
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–20. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

