Physical and data structure of 3D genome
- PMID: 31950084
- PMCID: PMC6954067
- DOI: 10.1126/sciadv.aay4055
Physical and data structure of 3D genome
Abstract
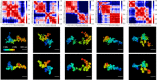
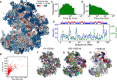
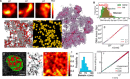
With the textbook view of chromatin folding based on the 30-nm fiber being challenged, it has been proposed that interphase DNA has an irregular 10-nm nucleosome polymer structure whose folding philosophy is unknown. Nevertheless, experimental advances suggest that this irregular packing is associated with many nontrivial physical properties that are puzzling from a polymer physics point of view. Here, we show that the reconciliation of these exotic properties necessitates modularizing three-dimensional genome into tree data structures on top of, and in striking contrast to, the linear topology of DNA double helix. These functional modules need to be connected and isolated by an open backbone that results in porous and heterogeneous packing in a quasi-self-similar manner, as revealed by our electron and optical imaging. Our multiscale theoretical and experimental results suggest the existence of higher-order universal folding principles for a disordered chromatin fiber to avoid entanglement and fulfill its biological functions.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures



References
-
- Maeshima K., Ide S., Babokhov M., Dynamic chromatin organization without the 30-Nm fiber. Curr. Opin. Cell Biol. 58, 95–104 (2019). - PubMed
-
- Almassalha L. M., Bauer G. M., Chandler J. E., Gladstein S., Cherkezyan L., Stypula-Cyrus Y., Weinberg S., Zhang D., Ruhoff P. T., Roy H. K., Subramanian H., Chandel N. S., Szleifer I., Backman V., Label-free imaging of the native, living cellular nanoarchitecture using partial-wave spectroscopic microscopy. Proc. Natl. Acad. Sci. U.S.A. 113, E6372–E6381 (2016). - PMC - PubMed
-
- Almassalha L. M., Bauer G. M., Wu W., Cherkezyan L., Zhang D., Kendra A., Gladstein S., Chandler J. E., VanDerway D., Seagle B.-L. L., Ugolkov A., Billadeau D. D., O’Halloran T. V., Mazar A. P., Roy H. K., Szleifer I., Shahabi S., Backman V., Macrogenomic engineering via modulation of the scaling of chromatin packing density. Nat. Biomed. Eng. 1, 902–913 (2017). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

