Heterochromatin formation in Drosophila requires genome-wide histone deacetylation in cleavage chromatin before mid-blastula transition in early embryogenesis
- PMID: 31950239
- PMCID: PMC7021753
- DOI: 10.1007/s00412-020-00732-x
Heterochromatin formation in Drosophila requires genome-wide histone deacetylation in cleavage chromatin before mid-blastula transition in early embryogenesis
Abstract
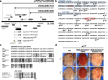
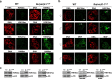
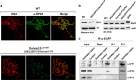
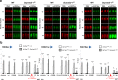
Su(var) mutations define epigenetic factors controlling heterochromatin formation and gene silencing in Drosophila. Here, we identify SU(VAR)2-1 as a novel chromatin regulator that directs global histone deacetylation during the transition of cleavage chromatin into somatic blastoderm chromatin in early embryogenesis. SU(VAR)2-1 is heterochromatin-associated in blastoderm nuclei but not in later stages of development. In larval polytene chromosomes, SU(VAR)2-1 is a band-specific protein. SU(VAR)2-1 directs global histone deacetylation by recruiting the histone deacetylase RPD3. In Su(var)2-1 mutants H3K9, H3K27, H4K8 and H4K16 acetylation shows elevated levels genome-wide and heterochromatin displays aberrant histone hyper-acetylation. Whereas H3K9me2- and HP1a-binding appears unaltered, the heterochromatin-specific H3K9me2S10ph composite mark is impaired in heterochromatic chromocenters of larval salivary polytene chromosomes. SU(VAR)2-1 contains an NRF1/EWG domain and a C2HC zinc-finger motif. Our study identifies SU(VAR)2-1 as a dosage-dependent, heterochromatin-initiating SU(VAR) factor, where the SU(VAR)2-1-mediated control of genome-wide histone deacetylation after cleavage and before mid-blastula transition (pre-MBT) is required to enable heterochromatin formation.
Keywords: Drosophila melanogaster; Heterochromatin; Histone deacetylation; Mid-blastula transition.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Baksa K, Morawietz H, Dombradi V, Axton M, Taubert H, Szabo G, Török I, Gyurkovics H, Szöör B, Gloover D, et al. Mutations in the phosphatase 1 gene at 87B can differentially affect suppression of position-effect variegation and mitosis in Drosophila melanogaster. Genetics. 1993;135:117–125. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

