Validation of the easyscreen flavivirus dengue alphavirus detection kit based on 3base amplification technology and its application to the 2016/17 Vanuatu dengue outbreak
- PMID: 31951602
- PMCID: PMC6968865
- DOI: 10.1371/journal.pone.0227550
Validation of the easyscreen flavivirus dengue alphavirus detection kit based on 3base amplification technology and its application to the 2016/17 Vanuatu dengue outbreak
Abstract
Background: The family flaviviridae and alphaviridae contain a diverse group of pathogens that cause significant morbidity and mortality worldwide. Diagnosis of the virus responsible for disease is essential to ensure patients receive appropriate clinical management. Very few real-time RT-PCR based assays are able to detect the presence of all members of these families using a single primer and probe set. We have developed a novel chemistry, 3base, which simplifies the viral nucleic acids allowing the design of RT-PCR assays capable of pan-family identification.
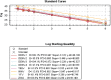
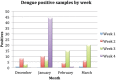
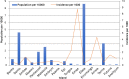
Methodology/principal finding: Synthetic constructs, viral nucleic acids, intact viral particles and characterised reference materials were used to determine the specificity and sensitivity of the assays. Synthetic constructs demonstrated the sensitivities of the pan-flavivirus detection component were in the range of 13 copies per PCR. The pan-alphavirus assay had a sensitivity range of 10-25 copies per reaction depending on the viral strain. Lower limit of detection studies using whole virus particles demonstrated that sensitivity for assays was in the range of 1-2 copies per reaction. No cross reactivity was observed with a number of commonly encountered viral strains. Proficiency panels showed 100% concordance with the expected results and the assays performed as well as, if not better than, other assays used in laboratories worldwide. After initial assay validation the pan-viral assays were then tested during the 2016-2017 Vanuatu dengue-2 outbreak. Positive results were detected in 116 positives from a total of 187 suspected dengue samples.
Conclusions/significance: The pan-viral screening assays described here utilise a novel 3base technology and are shown to provide a sensitive and specific method to screen and thereafter speciate flavi- and/or alpha- viruses in clinical samples. The assays performed well in an outbreak situation and can be used to detect positive clinical samples containing any flavivirus or alphavirus in approximately 3 hours 30 minutes.
Conflict of interest statement
Crystal Garae, Kalkoa Kalo and George Junior Pakoa have no competing interests. D. S. Millar, R. Baker and P. J. Isaacs are employees of Genetic Signatures and receive salary and equity interests in Genetic Signatures. This does not alter the authors' adherence to PLOS ONE policies on sharing data and materials.
Figures




References
-
- Shi P-Y, (editor) (2012). Molecular Virology and Control of Flaviviruses. Caister Academic Press. ISBN 978-1-904455-92-9
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

