Piece by piece: Building a ribozyme
- PMID: 31953324
- PMCID: PMC7039547
- DOI: 10.1074/jbc.REV119.009929
Piece by piece: Building a ribozyme
Abstract
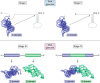
The ribosome and RNase P are cellular ribonucleoprotein complexes that perform peptide bond synthesis and phosphodiester bond cleavage, respectively. Both are ancient biological assemblies that were already present in the last universal common ancestor of all life. The large subunit rRNA in the ribosome and the RNA subunit of RNase P are the ribozyme components required for catalysis. Here, we explore the idea that these two large ribozymes may have begun their evolutionary odyssey as an assemblage of RNA "fragments" smaller than the contemporary full-length versions and that they transitioned through distinct stages along a pathway that may also be relevant for the evolution of other non-coding RNAs.
Keywords: enzyme catalysis; evolution; ribonuclease P (RNase P); ribonucleoprotein complex; ribosome; ribozyme (catalytic RNA) (RNA enzyme).
© 2020 Gray and Gopalan.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

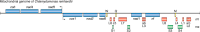

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

