Protein Engineering for Improving and Diversifying Natural Product Biosynthesis
- PMID: 31954530
- PMCID: PMC7274900
- DOI: 10.1016/j.tibtech.2019.12.008
Protein Engineering for Improving and Diversifying Natural Product Biosynthesis
Abstract
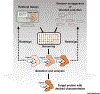
Proteins found in nature have traditionally been the most frequently used biocatalysts to produce numerous natural products ranging from commodity chemicals to pharmaceuticals. Protein engineering has emerged as a powerful biotechnological toolbox in the development of metabolic engineering, particularly for the biosynthesis of natural products. Recently, protein engineering has become a favored method to improve enzymatic activity, increase enzyme stability, and expand product spectra in natural product biosynthesis. This review summarizes recent advances and typical strategies in protein engineering, highlighting the paramount role of protein engineering in improving and diversifying the biosynthesis of natural products. Future prospects and research directions are also discussed.
Keywords: engineered biosynthesis; natural products; protein engineering.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Figures

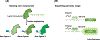

References
-
- Koehn FE and Carter GT (2005) The evolving role of natural products in drug discovery. Nature reviews Drug discovery 4 (3), 206. - PubMed
-
- Kole PL et al. (2005) Cosmetic potential of herbal extracts.
-
- Rodrigues T et al. (2016) Counting on natural products for drug design. Nat Chem 8 (6), 531–41. - PubMed
-
- O’Connor SE (2015) Engineering of Secondary Metabolism. Annu Rev Genet 49, 71–94. - PubMed

