Cellular Dialogues: Cell-Cell Communication through Diffusible Molecules Yields Dynamic Spatial Patterns
- PMID: 31954659
- PMCID: PMC6975168
- DOI: 10.1016/j.cels.2019.12.001
Cellular Dialogues: Cell-Cell Communication through Diffusible Molecules Yields Dynamic Spatial Patterns
Abstract
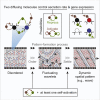
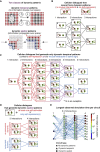
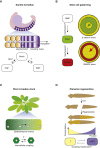
Cells form spatial patterns by coordinating their gene expressions. How a group of mesoscopic numbers (hundreds to thousands) of cells, without pre-existing morphogen gradients and spatial organization, self-organizes spatial patterns remains poorly understood. Of particular importance are dynamic spatial patterns such as spiral waves that perpetually move and transmit information. We developed an open-source software for simulating a field of cells that communicate by secreting any number of molecules. With this software and a theory, we identified all possible "cellular dialogues"-ways of communicating with two diffusing molecules-that yield diverse dynamic spatial patterns. These patterns emerge despite widely varying responses of cells to the molecules, gene-expression noise, spatial arrangements, and cell movements. A three-stage, "order-fluctuate-settle" process forms dynamic spatial patterns: cells form long-lived whirlpools of wavelets that, following erratic dynamics, settle into a dynamic spatial pattern. Our work helps in identifying gene-regulatory networks that underlie dynamic pattern formations.
Keywords: cell-cell communication; cellular automata; complex systems; gene networks; multicellular systems; pattern formation; reaction-diffusion; self organization; spatial patterns; waves.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures





References
-
- Alon U. First edition. CRC Press; 2006. An introduction to systems biology: design principles of biological circuits.
-
- Bar-Yam Y. First edition. Avalon Publishing; 2003. Dynamics of complex systems.
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

